Spatial and temporal analysis of the microbial community in slow sand filters used for treating horticultural irrigation water
- PMID: 12676691
- PMCID: PMC154832
- DOI: 10.1128/AEM.69.4.2116-2125.2003
Spatial and temporal analysis of the microbial community in slow sand filters used for treating horticultural irrigation water
Abstract
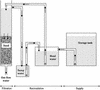
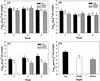
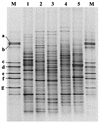
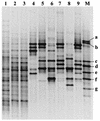
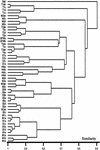
An experimental slow sand filter (SSF) was constructed to study the spatial and temporal structure of a bacterial community suppressive to an oomycete plant pathogen, Phytophthora cryptogea. Passage of water through the mature sand column resulted in complete removal of zoospores of the plant pathogen. To monitor global changes in the microbial community, bacterial and fungal numbers were estimated on selective media, direct viable counts of fungal spores were made, and the ATP content was measured. PCR amplification of 16S rRNA genes and denaturing gradient gel electrophoresis (DGGE) were used to study the dynamics of the bacterial community in detail. The top layer (1 cm) of the SSF column was dominated by a variable and active microbial population, whereas the middle (50 cm) and bottom (80 cm) layers were dominated by less active and diverse bacterial populations. The major changes in the microbial populations occurred during the first week of filter operation, and these populations then remained to the end of the study. Spatial and temporal nonlinear mapping of the DGGE bands provided a useful visual representation of the similarities between SSF samples. According to the DGGE profile, less than 2% of the dominating bands present in the SSF column were represented in the culturable population. Sequence analysis of DGGE bands from all depths of the SSF column indicated that a range of bacteria were present, with 16S rRNA gene sequences similar to groups such as Bacillus megaterium, Cytophaga, Desulfovibrio, Legionella, Rhodococcus rhodochrous, Sphingomonas, and an uncharacterized environmental clone. This study describes the characterization of the performance, and microbial composition, of SSFs used for the treatment of water for use in the horticultural industry. Utilization of naturally suppressive population of microorganisms either directly or by manipulation of the environment in an SSF may provide a more reproducible control method for the future.
Figures



References
-
- Ali-Shtayeh, M. S., and J. D. MacDonald. 1991. Occurrence of Phytophthora species in irrigation water in the Nablus area (West Bank of Jordan). Phytopathol. Mediterr. 30:143-150.
-
- Ali-Shtayeh, M. S., J. D. MacDonald, and J. Kabashima. 1991. A method for using commercial ELISA tests to detect zoospores of Phytophthora and Pythium species in irrigation water. Plant Dis. 75:305-311.
-
- Berkelmann, B., W. Wohanka, and G. Wolf. 1994. Characterisation of the bacterial flora in recirculating nutrient solutions of a hydroponic system with rockwool. Acta Hortic. 361:372-381.
-
- Bewley, W. F., and W. Buddin. 1921. On the fungus flora of glasshouse water supplies in relation to plant disease. Ann. Appl. Biol. 8:10-19.
-
- Bock, C., R. M. Kroppenstedt, and H. Diekmann. 1996. Degradation and bioconversion of aliphatic and aromatic hydrocarbons by Rhodococcus ruber 219. Appl. Microb. Biotechnol. 45:408-410.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

