Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome
- PMID: 12677556
- PMCID: PMC1180271
- DOI: 10.1086/375178
Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome
Abstract
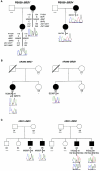
Bardet-Biedl syndrome is a genetically and clinically heterogeneous disorder caused by mutations in at least seven loci (BBS1-7), five of which are cloned (BBS1, BBS2, BBS4, BBS6, and BBS7). Genetic and mutational analyses have indicated that, in some families, a combination of three mutant alleles at two loci (triallelic inheritance) is necessary for pathogenesis. To date, four of the five known BBS loci have been implicated in this mode of oligogenic disease transmission. We present a comprehensive analysis of the spectrum, distribution, and involvement in non-Mendelian trait transmission of mutant alleles in BBS1, the most common BBS locus. Analyses of 259 independent families segregating a BBS phenotype indicate that BBS1 participates in complex inheritance and that, in different families, mutations in BBS1 can interact genetically with mutations at each of the other known BBS genes, as well as at unknown loci, to cause the phenotype. Consistent with this model, we identified homozygous M390R alleles, the most frequent BBS1 mutation, in asymptomatic individuals in two families. Moreover, our statistical analyses indicate that the prevalence of the M390R allele in the general population is consistent with an oligogenic rather than a recessive model of disease transmission. The distribution of BBS oligogenic alleles also indicates that all BBS loci might interact genetically with each other, but some genes, especially BBS2 and BBS6, are more likely to participate in triallelic inheritance, suggesting a variable ability of the BBS proteins to interact genetically with each other.
Figures



References
Electronic-Database Information
-
- BLAT search engine, http://genome.ucsc.edu/cgi-bin/hgBlat?command=start
-
- Genome Database, http://www.gdb.org
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BBS) - PubMed
-
- Whitehead Institute Center for Genome Research, http://www-genome.wi.mit.edu/
References
-
- Anderson KL, Lewis RA (1997) Bardet-Biedl Syndrome (BBS). J Rare Dis 3:5–10
-
- Badano JL, Katsanis N (2002) Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet 3:779–789 - PubMed
-
- Beales PL, Katsanis N, Lewis RA, Ansley SJ, Elcioglu N, Raza J, Woods MO, Green JS, Parfrey PS, Davidson WS, Lupski JR (2001) Genetic and mutational analyses of a large multiethnic Bardet-Biedl cohort reveal a minor involvement of BBS6 and delineate the critical intervals of other loci. Am J Hum Genet 68:606–616 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

