Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases
- PMID: 12679523
- PMCID: PMC153578
- DOI: 10.1073/pnas.0737647100
Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases
Abstract
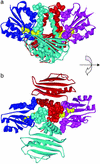
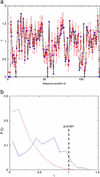
The binding between a PK and its target is highly specific, despite the fact that many different PKs exhibit significant sequence and structure homology. There must be, then, specificity-determining residues (SDRs) that enable different PKs to recognize their unique substrate. Here we use and further develop a computational procedure to discover putative SDRs (PSDRs) in protein families, whereby a family of homologous proteins is split into orthologous proteins, which are assumed to have the same specificity, and paralogous proteins, which have different specificities. We reason that PSDRs must be similar among orthologs, whereas they must necessarily be different among paralogs. Our statistical procedure and evolutionary model identifies such residues by discriminating a functional signal from a phylogenetic one. As case studies we investigate the prokaryotic two-component system and the eukaryotic AGC (i.e., cAMP-dependent PK, cGMP-dependent PK, and PKC) PKs. Without using experimental data, we predict PSDRs in prokaryotic and eukaryotic PKs, and suggest precise mutations that may convert the specificity of one PK to another. We compare our predictions with current experimental results and obtain considerable agreement with them. Our analysis unifies much of existing data on PK specificity. Finally, we find PSDRs that are outside the active site. Based on our results, as well as structural and biochemical characterizations of eukaryotic PKs, we propose the testable hypothesis of "specificity via differential activation" as a way for the cell to control kinase specificity.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

