Diversification and spectral tuning in marine proteorhodopsins
- PMID: 12682005
- PMCID: PMC154475
- DOI: 10.1093/emboj/cdg183
Diversification and spectral tuning in marine proteorhodopsins
Abstract
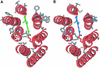
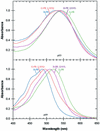
Proteorhodopsins, ubiquitous retinylidene photoactive proton pumps, were recently discovered in the cosmopolitan uncultured SAR86 bacterial group in oceanic surface waters. Two related proteorhodopsin families were found that absorb light with different absorption maxima, 525 nm (green) and 490 nm (blue), and their distribution was shown to be stratified with depth. Using structural modeling comparisons and mutagenesis, we report here on a single amino acid residue at position 105 that functions as a spectral tuning switch and accounts for most of the spectral difference between the two pigment families. Furthermore, looking at natural environments, we found novel proteorhodopsin gene clusters spanning the range of 540-505 nm and containing changes in the same identified key switch residue leading to changes in their absorption maxima. The results suggest a simultaneous diversification of green proteorhodopsin and the new key switch variant pigments. Our observations demonstrate that this single-residue switch mechanism is the major determinant of proteorhodopsin wavelength regulation in natural marine environments.
Figures



References
-
- Béjà O. et al. (2000a) Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science, 289, 1902–1906. - PubMed
-
- Béjà O. et al. (2000b) Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol., 2, 516–529. - PubMed
-
- Béjà O., Spudich,E.N., Spudich,J.L., Leclerc,M. and DeLong,E.F. (2001) Proteorhodopsin phototrophy in the ocean. Nature, 411, 786–789. - PubMed
-
- Bieszke J.A., Spudich,E.N., Scott,K.L., Borkovich,K.A. and Spudich,J.L. (1999) A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry, 38, 14138–14145. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

