The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane
- PMID: 12682011
- PMCID: PMC154466
- DOI: 10.1093/emboj/cdg174
The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane
Abstract
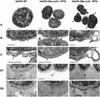
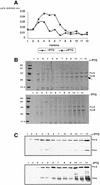
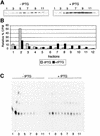
In Gram-negative bacteria, lipopolysaccharide and phospholipid biosynthesis takes place at the inner membrane. How the completed lipid molecules are subsequently transported to the outer membrane remains unknown. Omp85 of Neisseria meningitidis is representative for a family of outer membrane proteins conserved among Gram-negative bacteria. We first demonstrated that the omp85 gene is co-transcribed with genes involved in lipid biosynthesis, suggesting an involvement in lipid assembly. A meningococcal strain was constructed in which Omp85 expression could be switched on or off through a tac promoter-controlled omp85 gene. We demonstrated that the presence of Omp85 is essential for viability. Depletion of Omp85 leads to accumulation of electron-dense amorphous material and vesicular structures in the periplasm. We demonstrated, by fractionation of inner and outer membranes, that lipopolysaccharide and phospholipids mostly disappeared from the outer membrane and instead accumulated in the inner membrane, upon depletion of Omp85. Omp85 depletion did not affect localization of integral outer membrane proteins PorA and Opa. These results provide compelling evidence for a role for Omp85 in lipid transport to the outer membrane.
Figures




Similar articles
-
Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly.Res Microbiol. 2004 Apr;155(3):129-35. doi: 10.1016/j.resmic.2003.11.007. Res Microbiol. 2004. PMID: 15143770 Review.
-
Biogenesis of the Gram-negative bacterial outer membrane.Curr Opin Microbiol. 2004 Dec;7(6):610-6. doi: 10.1016/j.mib.2004.10.011. Curr Opin Microbiol. 2004. PMID: 15556033 Review.
-
Role of a highly conserved bacterial protein in outer membrane protein assembly.Science. 2003 Jan 10;299(5604):262-5. doi: 10.1126/science.1078973. Science. 2003. PMID: 12522254
-
Omp85 proteins of Neisseria gonorrhoeae and Neisseria meningitidis are similar to Haemophilus influenzae D-15-Ag and Pasteurella multocida Oma87.Microb Pathog. 1998 Jul;25(1):11-21. doi: 10.1006/mpat.1998.0206. Microb Pathog. 1998. PMID: 9705245
-
Molecular architecture and function of the Omp85 family of proteins.Mol Microbiol. 2005 Dec;58(5):1216-25. doi: 10.1111/j.1365-2958.2005.04906.x. Mol Microbiol. 2005. PMID: 16313611 Review.
Cited by
-
Highly conserved surface proteins of oral spirochetes as adhesins and potent inducers of proinflammatory and osteoclastogenic factors.Infect Immun. 2008 Jun;76(6):2428-38. doi: 10.1128/IAI.01128-07. Epub 2008 Apr 7. Infect Immun. 2008. PMID: 18390996 Free PMC article.
-
Drug Binding to BamA Targets Its Lateral Gate.J Phys Chem B. 2023 Aug 31;127(34):7509-7517. doi: 10.1021/acs.jpcb.3c04501. Epub 2023 Aug 16. J Phys Chem B. 2023. PMID: 37587651 Free PMC article.
-
Targeting BAM for Novel Therapeutics against Pathogenic Gram-Negative Bacteria.Antibiotics (Basel). 2023 Mar 30;12(4):679. doi: 10.3390/antibiotics12040679. Antibiotics (Basel). 2023. PMID: 37107041 Free PMC article. Review.
-
Outer membrane protein biogenesis in Gram-negative bacteria.Philos Trans R Soc Lond B Biol Sci. 2015 Oct 5;370(1679):20150023. doi: 10.1098/rstb.2015.0023. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26370935 Free PMC article. Review.
-
Evidence for conservation of architecture and physical properties of Omp85-like proteins throughout evolution.Proc Natl Acad Sci U S A. 2004 Oct 5;101(40):14497-502. doi: 10.1073/pnas.0404679101. Epub 2004 Sep 20. Proc Natl Acad Sci U S A. 2004. PMID: 15381771 Free PMC article.
References
-
- Ainsworth S.K., Ito,S. and Karnovsky,M.J. (1972) Alkaline bismuth reagent for high resolution ultrastructural demonstration of periodate-reactive sites. J. Histochem. Cytochem., 20, 995–1005. - PubMed
-
- Apicella M.A., Griffiss,J.M. and Schneider,H. (1994) Isolation and characterization of lipopolysaccharides, lipooligosaccharides and lipid A. Methods Enzymol., 235, 242–252. - PubMed
-
- Bayer M.E. (1968) Areas of adhesion between wall and membrane of Escherichia coli. J. Gen. Microbiol., 53, 395–404. - PubMed
-
- Bligh E.G. and Dyer,W.J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol., 37, 911–917. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

