Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm
- PMID: 12682020
- PMCID: PMC154478
- DOI: 10.1093/emboj/cdg187
Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm
Abstract
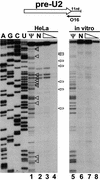
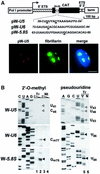
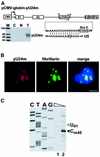
Biogenesis of functional spliceosomal small nuclear RNAs (snRNAs) includes the post-transcriptional covalent modification of numerous internal nucleotides. We have recently demonstrated that synthesis of 2'-O-methylated nucleotides and pseudouridines in the RNA polymerase II-synthesized Sm snRNAs is directed by sequence-specific guide RNAs. Here, we provide evidence supporting the notion that modification of Sm snRNAs occurs in nucleoplasmic Cajal bodies (CBs), where modification guide RNAs accumulate. We show that short fragments of Sm snRNAs are correctly and efficiently modified when targeted to CBs, but not when these same fragments are targeted to the nucleolus. We also demonstrate that internal modification of the U2 snRNA occurs exclusively after nuclear import of the newly assembled Sm snRNP from the cytoplasm. Finally, we show that p80 coilin, the CB marker protein, is not required for snRNA modification. In coilin knockout cells, Sm snRNAs and their modification guide RNAs colocalize in residual CBs, which do not stockpile fibrillarin and fail to recruit the U3 small nucleolar RNA.
Figures
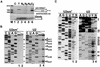

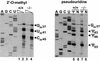
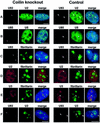
Similar articles
-
A role for Cajal bodies in the final steps of U2 snRNP biogenesis.J Cell Sci. 2004 Sep 1;117(Pt 19):4423-33. doi: 10.1242/jcs.01308. Epub 2004 Aug 17. J Cell Sci. 2004. PMID: 15316075
-
Cajal body-specific small nuclear RNAs: a novel class of 2'-O-methylation and pseudouridylation guide RNAs.EMBO J. 2002 Jun 3;21(11):2746-56. doi: 10.1093/emboj/21.11.2746. EMBO J. 2002. PMID: 12032087 Free PMC article.
-
Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product.J Cell Biol. 2001 Jul 23;154(2):293-307. doi: 10.1083/jcb.200104083. J Cell Biol. 2001. PMID: 11470819 Free PMC article.
-
The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze.Chromosoma. 2006 Oct;115(5):343-54. doi: 10.1007/s00412-006-0056-6. Epub 2006 Mar 31. Chromosoma. 2006. PMID: 16575476 Review.
-
Activation of transcription enforces the formation of distinct nuclear bodies in zebrafish embryos.RNA Biol. 2017 Jun 3;14(6):752-760. doi: 10.1080/15476286.2016.1255397. Epub 2016 Nov 18. RNA Biol. 2017. PMID: 27858508 Free PMC article. Review.
Cited by
-
CRM1 plays a nuclear role in transporting snoRNPs to nucleoli in higher eukaryotes.Nucleus. 2012 Mar 1;3(2):132-7. doi: 10.4161/nucl.19266. Epub 2012 Mar 1. Nucleus. 2012. PMID: 22555597 Free PMC article. Review.
-
Specific genomic cues regulate Cajal body assembly.RNA Biol. 2017 Jun 3;14(6):791-803. doi: 10.1080/15476286.2016.1243648. Epub 2016 Oct 7. RNA Biol. 2017. PMID: 27715441 Free PMC article. Review.
-
Detection and quantitation of RNA base modifications.RNA. 2004 Jun;10(6):996-1002. doi: 10.1261/rna.7110804. RNA. 2004. PMID: 15146083 Free PMC article.
-
Identification of 13 novel human modification guide RNAs.Nucleic Acids Res. 2003 Nov 15;31(22):6543-51. doi: 10.1093/nar/gkg849. Nucleic Acids Res. 2003. PMID: 14602913 Free PMC article.
-
A day in the life of the spliceosome.Nat Rev Mol Cell Biol. 2014 Feb;15(2):108-21. doi: 10.1038/nrm3742. Nat Rev Mol Cell Biol. 2014. PMID: 24452469 Free PMC article. Review.
References
-
- Bakin A. and Ofengand,J. (1993) Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry, 32, 9754–9762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

