Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning
- PMID: 12682023
- PMCID: PMC154467
- DOI: 10.1093/emboj/cdg175
Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning
Abstract
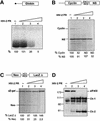
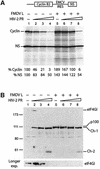
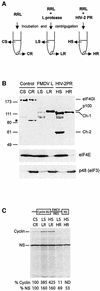
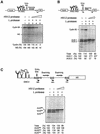
The eukaryotic translation initiation factor eIF4GI binds several proteins and acts as a scaffold to promote preinitiation complex formation on the mRNA molecule (48S). Following mRNA attachment this complex scans along the messenger in a 5' to 3' direction until it locates and recognizes the initiation start codon. By using a combination of retroviral and picornaviral proteases (HIV-2 and L respectively) in the reticulocyte lysate system, we have characterized a 40 amino acid (aa) region of eIF4GI (aa 642-681) that exhibits general RNA-binding properties. Removal of this domain by proteolytic processing followed by translational assays showed virtually no inhibition of internal ribosome entry on the encephalomyocarditis virus, but resulted in drastic impairment of ribosome scanning as demonstrated by studying poliovirus and foot-and-mouth disease virus translation. Based on these findings, we propose that this 40 aa motif of eIF4GI is critical for ribosome scanning.
Figures





Similar articles
-
In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system.J Mol Biol. 2002 Apr 19;318(1):9-20. doi: 10.1016/S0022-2836(02)00070-0. J Mol Biol. 2002. PMID: 12054764
-
Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region.Mol Cell Biol. 2000 Jan;20(2):468-77. doi: 10.1128/MCB.20.2.468-477.2000. Mol Cell Biol. 2000. PMID: 10611225 Free PMC article.
-
Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation.Mol Cell Biol. 2001 Jul;21(13):4097-109. doi: 10.1128/MCB.21.13.4097-4109.2001. Mol Cell Biol. 2001. PMID: 11390639 Free PMC article.
-
Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites.C R Biol. 2005 Jul;328(7):589-605. doi: 10.1016/j.crvi.2005.02.004. C R Biol. 2005. PMID: 15992743 Review.
-
ABCE1: A special factor that orchestrates translation at the crossroad between recycling and initiation.RNA Biol. 2017 Oct 3;14(10):1279-1285. doi: 10.1080/15476286.2016.1269993. Epub 2017 May 12. RNA Biol. 2017. PMID: 28498001 Free PMC article. Review.
Cited by
-
A mechanistic overview of translation initiation in eukaryotes.Nat Struct Mol Biol. 2012 Jun 5;19(6):568-76. doi: 10.1038/nsmb.2303. Nat Struct Mol Biol. 2012. PMID: 22664984 Review.
-
miRNA repression of translation in vitro takes place during 43S ribosomal scanning.Nucleic Acids Res. 2013 Jan 7;41(1):586-98. doi: 10.1093/nar/gks1076. Epub 2012 Nov 17. Nucleic Acids Res. 2013. PMID: 23161679 Free PMC article.
-
Poliovirus 2A(Pro) increases viral mRNA and polysome stability coordinately in time with cleavage of eIF4G.J Virol. 2008 Jun;82(12):5847-59. doi: 10.1128/JVI.01514-07. Epub 2008 Apr 9. J Virol. 2008. PMID: 18400852 Free PMC article.
-
The role of the poly(A) binding protein in the assembly of the Cap-binding complex during translation initiation in plants.Translation (Austin). 2014 Oct 30;2(2):e959378. doi: 10.4161/2169074X.2014.959378. eCollection 2014 Sep 1. Translation (Austin). 2014. PMID: 26779409 Free PMC article. Review.
-
Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation.Nucleic Acids Res. 2007;35(18):e121. doi: 10.1093/nar/gkm682. Epub 2007 Sep 18. Nucleic Acids Res. 2007. PMID: 17881372 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

