Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2
- PMID: 12682024
- PMCID: PMC154477
- DOI: 10.1093/emboj/cdg185
Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2
Abstract
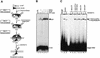
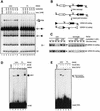
The RAG1 and RAG2 proteins perform critical DNA recognition and cleavage functions in V(D)J recombination, and also catalyze efficient DNA transposition in vitro. No transposition in vivo by the RAG proteins has been reported, suggesting regulation of the reaction by as yet unknown mechanisms. Here we report that RAG-mediated transposition is suppressed by physiological concentrations of the guanine nucleotide GTP, and by the full-length RAG2 protein. Both GTP and full-length RAG2 inhibit transposition by blocking the non-covalent 'capture' of target DNA, and both are capable of inhibiting RAG-mediated hybrid joint formation in vitro. We also observe that another intracellular signaling molecule, Ca(2+), stimulates RAG-mediated transposition and is capable of activating transposition even in reactions containing full-length RAG2 and GTP. RAG-mediated transposition has been proposed to contribute to the chromosomal translocations that underlie the development of lymphoid malignancies, and our findings highlight regulatory mechanisms that might prevent such occurrences, and circumstances in which these regulatory mechanisms could be overcome.
Figures


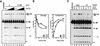
Similar articles
-
The C-terminal portion of RAG2 protects against transposition in vitro.EMBO J. 2003 Apr 15;22(8):1931-8. doi: 10.1093/emboj/cdg184. EMBO J. 2003. PMID: 12682025 Free PMC article.
-
New concepts in the regulation of an ancient reaction: transposition by RAG1/RAG2.Immunol Rev. 2004 Aug;200:261-71. doi: 10.1111/j.0105-2896.2004.00167.x. Immunol Rev. 2004. PMID: 15242411 Review.
-
RAG1 and RAG2 in V(D)J recombination and transposition.Immunol Res. 2001;23(1):23-39. doi: 10.1385/IR:23:1:23. Immunol Res. 2001. PMID: 11417858 Review.
-
The RAG proteins and V(D)J recombination: complexes, ends, and transposition.Annu Rev Immunol. 2000;18:495-527. doi: 10.1146/annurev.immunol.18.1.495. Annu Rev Immunol. 2000. PMID: 10837067 Review.
-
Mobilization of RAG-generated signal ends by transposition and insertion in vivo.Mol Cell Biol. 2006 Feb;26(4):1558-68. doi: 10.1128/MCB.26.4.1558-1568.2006. Mol Cell Biol. 2006. PMID: 16449665 Free PMC article.
Cited by
-
In vivo reinsertion of excised episomes by the V(D)J recombinase: a potential threat to genomic stability.PLoS Biol. 2007 Mar;5(3):e43. doi: 10.1371/journal.pbio.0050043. PLoS Biol. 2007. PMID: 17298184 Free PMC article.
-
Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination.Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22487-92. doi: 10.1073/pnas.1014958107. Epub 2010 Dec 13. Proc Natl Acad Sci U S A. 2010. PMID: 21149691 Free PMC article.
-
RAG: a recombinase diversified.Nat Immunol. 2009 Aug;10(8):817-21. doi: 10.1038/ni.1776. Epub 2009 Jul 21. Nat Immunol. 2009. PMID: 19621044 Free PMC article. Review.
-
Genetic evidence that GTP is required for transposition of IS903 and Tn552 in Escherichia coli.J Bacteriol. 2005 Jul;187(13):4598-606. doi: 10.1128/JB.187.13.4598-4606.2005. J Bacteriol. 2005. PMID: 15968071 Free PMC article.
-
Autoubiquitylation of the V(D)J recombinase protein RAG1.Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15446-51. doi: 10.1073/pnas.2637012100. Epub 2003 Dec 11. Proc Natl Acad Sci U S A. 2003. PMID: 14671314 Free PMC article.
References
-
- Agrawal A. and Schatz,D.G. (1997) RAG1 and RAG2 form a stable post-cleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell, 89, 43–53. - PubMed
-
- Agrawal A., Eastman,Q.M. and Schatz,D.G. (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature, 394, 744–751. - PubMed
-
- Aidinis V., Dias,D.C., Gomez,C.A., Bhattacharyya,D., Spanopoulou,E. and Santagata,S. (2000) Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol., 164, 5826–5832. - PubMed
-
- Alberts B., Bray,D., Lewis,J., Raff,M., Roberts,K. and Watson,J.D. (1994) Molecular Biology of the Cell. Garland Publishing, Inc., New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

