The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus
- PMID: 12682108
- PMCID: PMC2193890
- DOI: 10.1084/jem.20021417
The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus
Abstract
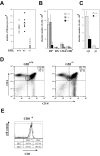
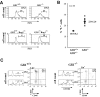
In the thymus, several steps of proliferative expansion and selection coordinate the maturation of precursors into antigen-specific T cells. Here we identify the transcriptional repressor Gfi1 as an important regulator of this maturation process. Mice lacking Gfi1 show reduced thymic cellularity due to an increased cell death rate, lack of proliferation, and a differentiation block in the very early uncommitted CD4-/CD8-/c-Kit+ cytokine-dependent T cell progenitors that have not yet initiated VDJ recombination. In addition, Gfi1-deficient mice show increased major histocompatibility complex class I-restricted positive selection and develop significantly more CD8+ cells suggesting a requirement of Gfi1 for a correct CD4/CD8 lineage decision. Absence of Gfi1 correlates with high level expression of the genes for lung Krüppel-like factor (LKLF), inhibitor of DNA binding (Id)1 and Id2, suggesting the existence of new regulatory pathways in pre-T cell development and thymic selection in which Gfi1 acts upstream of LKLF as well as the E-proteins, which are negatively regulated by Id1 and Id2.
Figures





Similar articles
-
Differential impact of the transcriptional repressor Gfi1 on mature CD4+ and CD8+ T lymphocyte function.Eur J Immunol. 2007 Dec;37(12):3551-63. doi: 10.1002/eji.200737130. Eur J Immunol. 2007. PMID: 18034420
-
Phenotypic and functional characterization of c-kit expression during intrathymic T cell development.J Immunol. 1992 Oct 1;149(7):2281-5. J Immunol. 1992. PMID: 1382094
-
CD2-CD4-CD8- lymph node T lymphocytes in MRL lpr/lpr mice are derived from a CD2+CD4+CD8+ thymic precursor.J Immunol. 1993 Jul 15;151(2):1086-96. J Immunol. 1993. PMID: 7687614
-
CD4 and CD8 act as co-receptors during thymic selection of the T cell repertoire.Semin Immunol. 1991 May;3(3):167-75. Semin Immunol. 1991. PMID: 1909594 Review.
-
A positive look at double-negative thymocytes.Nat Rev Immunol. 2002 Nov;2(11):888-97. doi: 10.1038/nri937. Nat Rev Immunol. 2002. PMID: 12415312 Review.
Cited by
-
Targeting tetramer-forming GABPβ isoforms impairs self-renewal of hematopoietic and leukemic stem cells.Cell Stem Cell. 2012 Aug 3;11(2):207-19. doi: 10.1016/j.stem.2012.05.021. Cell Stem Cell. 2012. PMID: 22862946 Free PMC article.
-
Transcription factors and target genes of pre-TCR signaling.Cell Mol Life Sci. 2015 Jun;72(12):2305-21. doi: 10.1007/s00018-015-1864-8. Epub 2015 Feb 22. Cell Mol Life Sci. 2015. PMID: 25702312 Free PMC article. Review.
-
Gfi-1B promoter remains associated with active chromatin marks throughout erythroid differentiation of human primary progenitor cells.Stem Cells. 2009 Sep;27(9):2153-62. doi: 10.1002/stem.151. Stem Cells. 2009. PMID: 19522008 Free PMC article.
-
From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation.Blood. 2015 Dec 10;126(24):2561-9. doi: 10.1182/blood-2015-06-655043. Epub 2015 Oct 7. Blood. 2015. PMID: 26447191 Free PMC article. Review.
-
The growth factor independence-1 transcription factor: new functions and new insights.Crit Rev Oncol Hematol. 2006 Aug;59(2):85-97. doi: 10.1016/j.critrevonc.2006.02.002. Epub 2006 May 23. Crit Rev Oncol Hematol. 2006. PMID: 16716599 Free PMC article. Review.
References
-
- Rödel, B., T. Wagner, M. Zörnig, J. Niessing, and T. Möröy. 1998. The human homologue (GFI1B) of the chicken GFI gene maps to chromosome 9q34.13-A locus frequently altered in hematopoietic diseases. Genomics. 54:580–582. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

