Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing
- PMID: 12682154
- PMCID: PMC153921
- DOI: 10.1128/JCM.41.4.1623-1636.2003
Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing
Abstract
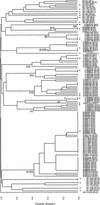
A multilocus sequence typing (MLST) scheme has been developed for the unambiguous characterization of encapsulated and noncapsulated Haemophilus influenzae isolates. The sequences of internal fragments of seven housekeeping genes were determined for 131 isolates, comprising a diverse set of 104 serotype a, b, c, d, e, and f isolates and 27 noncapsulated isolates. Many of the encapsulated isolates had previously been characterized by multilocus enzyme electrophoresis (MLEE), and the validity of the MLST scheme was established by the very similar clustering of isolates obtained by these methods. Isolates of serotypes c, d, e, and f formed monophyletic groups on a dendrogram constructed from the differences in the allelic profiles of the isolates, whereas there were highly divergent lineages of both serotype a and b isolates. Noncapsulated isolates were distinct from encapsulated isolates and, with one exception, were within two highly divergent clusters. The relationships between the major lineages of encapsulated H. influenzae inferred from MLEE data could not be discerned on a dendrogram constructed from differences in the allelic profiles, but were apparent on a tree reconstructed from the concatenated nucleotide sequences. Recombination has not therefore completely eliminated phylogenetic signal, and in support of this, for encapsulated isolates, there was significant congruence between many of the trees reconstructed from the sequences of the seven individual loci. Congruence was less apparent for noncapsulated isolates, suggesting that the impact of recombination is greater among noncapsulated than encapsulated isolates. The H. influenzae MLST scheme is available at www.mlst.net, it allows any isolate to be compared with those in the MLST database, and (for encapsulated isolates) it assigns isolates to their phylogenetic lineage, via the Internet.
Figures


Similar articles
-
Identification of Haemophilus influenzae serotype e strains missing the fucK gene in clinical isolates from Japan.J Med Microbiol. 2019 Oct;68(10):1534-1539. doi: 10.1099/jmm.0.001055. J Med Microbiol. 2019. PMID: 31368885
-
Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands.J Clin Microbiol. 2005 Jun;43(6):2741-9. doi: 10.1128/JCM.43.6.2741-2749.2005. J Clin Microbiol. 2005. PMID: 15956392 Free PMC article.
-
Emergence of nonencapsulated and encapsulated non-b-type invasive Haemophilus influenzae isolates in Portugal (1989-2001).J Clin Microbiol. 2004 Feb;42(2):807-10. doi: 10.1128/JCM.42.2.807-810.2004. J Clin Microbiol. 2004. PMID: 14766857 Free PMC article.
-
Molecular epidemiology and evolution of Haemophilus influenzae.Infect Genet Evol. 2020 Jun;80:104205. doi: 10.1016/j.meegid.2020.104205. Epub 2020 Jan 22. Infect Genet Evol. 2020. PMID: 31981610 Review.
-
Haemophilus influenzae serotype a as a cause of serious invasive infections.Lancet Infect Dis. 2014 Jan;14(1):70-82. doi: 10.1016/S1473-3099(13)70170-1. Epub 2013 Nov 20. Lancet Infect Dis. 2014. PMID: 24268829 Review.
Cited by
-
Haemophilus influenzae one day in Denmark: prevalence, circulating clones, and dismal resistance to aminopenicillins.Eur J Clin Microbiol Infect Dis. 2021 Oct;40(10):2077-2085. doi: 10.1007/s10096-021-04247-w. Epub 2021 Apr 23. Eur J Clin Microbiol Infect Dis. 2021. PMID: 33891188
-
Population subdivision and the detection of recombination in non-typable Haemophilus influenzae.Microbiology (Reading). 2012 Dec;158(Pt 12):2958-2964. doi: 10.1099/mic.0.063073-0. Epub 2012 Oct 4. Microbiology (Reading). 2012. PMID: 23038806 Free PMC article.
-
Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains.Genome Biol. 2007;8(6):R103. doi: 10.1186/gb-2007-8-6-r103. Genome Biol. 2007. PMID: 17550610 Free PMC article.
-
Characteristics of Haemophilus influenzae type b responsible for meningitis in Poland from 1997 to 2004.J Clin Microbiol. 2005 Nov;43(11):5665-9. doi: 10.1128/JCM.43.11.5665-5669.2005. J Clin Microbiol. 2005. PMID: 16272502 Free PMC article.
-
Molecular epidemiology and antimicrobial resistance of Haemophilus influenzae in Guiyang, Guizhou, China.Front Public Health. 2022 Dec 1;10:947051. doi: 10.3389/fpubh.2022.947051. eCollection 2022. Front Public Health. 2022. PMID: 36530676 Free PMC article.
References
-
- Anonymous. 2002. Progress toward elimination of Haemophilus influenzae type b invasive disease among infants and children—United States, 1998-2000. Morb. Mortal. Wkly. Rep. 51:234-237. - PubMed
-
- Breukels, M. A., L. Spanjaard. L.A. Sanders, and G. T. Rijkers. 2001. Immunological characterization of conjugated Haemophilus influenzae type b vaccine failure in infants. Clin. Infect. Dis. 32:1700-1705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

