BIBI, a bioinformatics bacterial identification tool
- PMID: 12682188
- PMCID: PMC153906
- DOI: 10.1128/JCM.41.4.1785-1787.2003
BIBI, a bioinformatics bacterial identification tool
Abstract
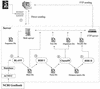
BIBI was designed to automate DNA sequence analysis for bacterial identification in the clinical field. BIBI relies on the use of BLAST and CLUSTAL W programs applied to different subsets of sequences extracted from GenBank. These sequences are filtered and stored in a new database, which is adapted to bacterial identification.
Figures

References
-
- Kolbert, C. P., and D. H. Persing. 1999. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr. Opin. Microbiol. 2:299-305. - PubMed
-
- Morgenstern, B., K. Frech, A. Dress, and T. Werner. 1998. DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics 14:290-294. - PubMed
-
- Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

