GenDB--an open source genome annotation system for prokaryote genomes
- PMID: 12682369
- PMCID: PMC153740
- DOI: 10.1093/nar/gkg312
GenDB--an open source genome annotation system for prokaryote genomes
Abstract
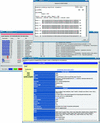
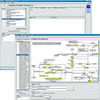
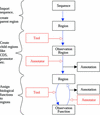
The flood of sequence data resulting from the large number of current genome projects has increased the need for a flexible, open source genome annotation system, which so far has not existed. To account for the individual needs of different projects, such a system should be modular and easily extensible. We present a genome annotation system for prokaryote genomes, which is well tested and readily adaptable to different tasks. The modular system was developed using an object-oriented approach, and it relies on a relational database backend. Using a well defined application programmers interface (API), the system can be linked easily to other systems. GenDB supports manual as well as automatic annotation strategies. The software currently is in use in more than a dozen microbial genome annotation projects. In addition to its use as a production genome annotation system, it can be employed as a flexible framework for the large-scale evaluation of different annotation strategies. The system is open source.
Figures

References
-
- Gaasterland T. and Sensen,C.W. (1996) MAGPIE: automated genome interpretation. Trends Genet., 12, 76–78. - PubMed
-
- Andrade M.A., Brown,N.P., Leroy,C., Hoersch,S., de Daruvar,A., Reich,C., Franchini,A., Tamames,J., Valencia,A., Ouzounis,C. and Sander,C. (1999) Automated genome sequence analysis and annotation. Bioinformatics, 15, 391–412. - PubMed
-
- Frishman D., Albermann,K., Hani,J., Heumann,K., Metanomski,A., Zollner,A. and Mewes,H.W. (2001) Functional and structural genomics using PEDANT. Bioinformatics, 17, 44–57. - PubMed
-
- Rutherford K.M., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.-A. and Barrell,B. (2000) Artemis: sequence visualisation and annotation. Bioinformatics, 16, 944–945. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

