Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways
- PMID: 12686616
- PMCID: PMC153129
- DOI: 10.1091/mbc.e02-08-0456
Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways
Abstract
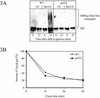
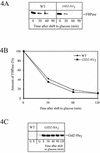
Metabolic adaptation of Saccharomyces cerevisiae cells from a nonfermentable carbon source to glucose induces selective, rapid breakdown of the gluconeogenetic key enzyme fructose-1,6-bisphosphatase (FBPase), a process called catabolite degradation. Herein, we identify eight novel GID genes required for proteasome-dependent catabolite degradation of FBPase. Four yeast proteins contain the CTLH domain of unknown function. All of them are Gid proteins. The site of catabolite degradation has been controversial until now. Two FBPase degradation pathways have been described, one dependent on the cytosolic ubiquitin-proteasome machinery, and the other dependent on vacuolar proteolysis. Interestingly, three of the novel Gid proteins involved in ubiquitin-proteasome-dependent degradation have also been reported by others to affect the vacuolar degradation pathway. As shown herein, additional genes suggested to be essential for vacuolar degradation are unnecessary for proteasome-dependent degradation. These data raise the question as to whether two FBPase degradation pathways exist that share components. Detailed characterization of Gid2p demonstrates that it is part of a soluble, cytosolic protein complex of at least 600 kDa. Gid2p is necessary for FBPase ubiquitination. Our studies have not revealed any involvement of vesicular intermediates in proteasome-dependent FBPase degradation. The influence of Ubp14p, a deubiquitinating enzyme, on proteasome-dependent catabolite degradation was further uncovered.
Figures






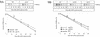
References
-
- Ausubel FM, Kingston RE, Seidman FG, Struhl K, Moore DD, Brent R, Smith FA. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1992.
-
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–86. - PubMed
-
- Barth H, Thumm M. A genomic screen identifies AUT8 as a novel gene essential for autophagy in the yeast Saccharomyces cerevisiae. Gene. 2001;274:151–156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

