Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma
- PMID: 12687498
- PMCID: PMC1180267
- DOI: 10.1086/375034
Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma
Abstract
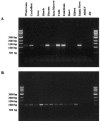
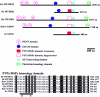
Charcot-Marie-Tooth disease (CMT) with autosomal recessive (AR) inheritance is a heterogeneous group of inherited motor and sensory neuropathies. In some families from Japan and Brazil, a demyelinating CMT, mainly characterized by the presence of myelin outfoldings on nerve biopsies, cosegregated as an autosomal recessive trait with early-onset glaucoma. We identified two such large consanguineous families from Tunisia and Morocco with ages at onset ranging from 2 to 15 years. We mapped this syndrome to chromosome 11p15, in a 4.6-cM region overlapping the locus for an isolated demyelinating ARCMT (CMT4B2). In these two families, we identified two different nonsense mutations in the myotubularin-related 13 gene, MTMR13. The MTMR protein family includes proteins with a phosphoinositide phosphatase activity, as well as proteins in which key catalytic residues are missing and that are thus called "pseudophosphatases." MTM1, the first identified member of this family, and MTMR2 are responsible for X-linked myotubular myopathy and Charcot-Marie-Tooth disease type 4B1, an isolated peripheral neuropathy with myelin outfoldings, respectively. Both encode active phosphatases. It is striking to note that mutations in MTMR13 also cause peripheral neuropathy with myelin outfoldings, although it belongs to a pseudophosphatase subgroup, since its closest homologue is MTMR5/Sbf1. This is the first human disease caused by mutation in a pseudophosphatase, emphasizing the important function of these putatively inactive enzymes. MTMR13 may be important for the development of both the peripheral nerves and the trabeculum meshwork, which permits the outflow of the aqueous humor. Both of these tissues have the same embryonic origin.
Figures


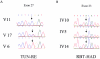

References
Electronic-Database Information
-
- Center for Medical Genetics, Marshfield Clinic, http://research.marshfieldclinic.org/genetics/
-
- Ensembl Genome Browser, http://www.ensembl.org/ (for Ensembl database)
-
- Entrez-Protein, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=protein (for hMTM1 [accession number Q13496], hMTMR1 [accession number Q13613], hMTMR2 [accession number Q13614], hMTMR3 [accession number Q13615], hMTMR4 [accession number AAH35609], hMTMR5 [accession number O95248], hMTMR6 [accession number Q9Y217], hMTMR7 [accession number Q9Y216], hMTMR8 [accession number AAH12399], hMTMR9 [accession number CAC51114], hMTMR10 [accession number AK000320], hMTMR11 [accession number AY028703], ScMTM [accession number P47147], DmCG3497 [accession number AAF48390], DmCG3530 [accession number AAF46997], DmCG3632 [accession number AAF48581], DmCG6939 [accession number AAK93570], DmCG3553 [accession number AAF50343], and DmCG9115 [accession number AAF52327])
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for LOC196111 [accession number XM_113650], LOC283105 [accession number XM_208513], KIAA1766 [accession number XM_049218], Agencourt_6446961 [accession number BM803570, contig [accession number NT_028309.7], and genomic clones [accession numbers AC080023.6, AC100763.2, AC011092.4, and AC026250.16])
-
- Genome Database, http://www.gdb.org/
References
-
- Akarsu AN, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS, Sarfarazi M (1996) A second locus (GLC3B) for primary congenital glaucoma (buphthalmos) maps to the 1p36 region. Hum Mol Genet 5:1199–1203 - PubMed
-
- Arruda WO, Comerlato EA, Scola RH, Silvado CE, Werneck LC (1999) Hereditary motor and sensory neuropathy with congenital glaucoma. Report on a family. Arq Neuropsiquiatr 57:190–194 - PubMed
-
- Baxter RV, Ben Othmane K, Rochelle JM, Stajich JE, Hulette C, Dew-Knight S, Hentati F, Ben Hamida M, Bel S, Stenger JE, Gilbert JR, Pericak-Vance MA, Vance JM (2002) Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat Genet 30:21–22 - PubMed
-
- Ben Othmane K, Hentati F, Lennon F, Ben Hamida C, Blel S, Roses AD, Pericak-Vance MA, Ben Hamida M, Vance JM (1993) Linkage of a locus (CMT4A) for autosomal recessive Charcot-Marie-Tooth disease to chromosome 8q. Hum Mol Genet 2:1625–1628 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

