Analysis of 4.3 kilobases of divergent locus B of macaque retroperitoneal fibromatosis-associated herpesvirus reveals a close similarity in gene sequence and genome organization to Kaposi's sarcoma-associated herpesvirus
- PMID: 12692211
- PMCID: PMC153986
- DOI: 10.1128/jvi.77.9.5084-5097.2003
Analysis of 4.3 kilobases of divergent locus B of macaque retroperitoneal fibromatosis-associated herpesvirus reveals a close similarity in gene sequence and genome organization to Kaposi's sarcoma-associated herpesvirus
Abstract
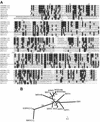
We previously identified retroperitoneal fibromatosis-associated herpesvirus (RFHV) as a simian homolog of Kaposi's sarcoma-associated herpesvirus (KSHV) in a fibroproliferative malignancy of macaques that has similarities to Kaposi's sarcoma. In this report, we cloned 4.3 kb of divergent locus B (DL-B) flanking the DNA polymerase gene from two variants of RFHV from different species of macaque with a consensus degenerate hybrid oligonucleotide primer approach. Within the DL-B region of RFHV, viral homologs of the cellular interleukin-6, dihydrofolate reductase, and thymidylate synthase genes were identified, along with a homolog of the gammaherpesvirus open reading frame (ORF) 10. In addition, a homolog of the KSHV ORF K3, the modulator of immune recognition-1, was identified. Our data show a close similarity in sequence conservation, gene content, and genomic structure between RFHV and KSHV which strongly supports the grouping of these viral species within the same RV-1 rhadinovirus lineage and the hypothesis that RFHV is the macaque homolog of KSHV.
Figures










References
-
- Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. - PubMed
-
- Burger, R., F. Neipel, B. Fleckenstein, R. Savino, G. Ciliberto, J. R. Kalden, and M. Gramatzki. 1998. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cells. Blood 91:1858-1863. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

