Mutations in the N-terminal region of influenza virus PB2 protein affect virus RNA replication but not transcription
- PMID: 12692212
- PMCID: PMC153989
- DOI: 10.1128/jvi.77.9.5098-5108.2003
Mutations in the N-terminal region of influenza virus PB2 protein affect virus RNA replication but not transcription
Abstract
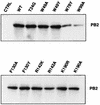
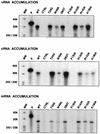
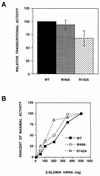
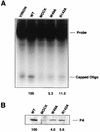
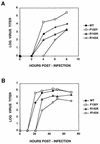
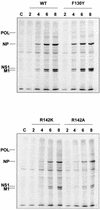
PB2 mutants of influenza virus were prepared by altering conserved positions in the N-terminal region of the protein that aligned with the amino acids of the eIF4E protein, involved in cap recognition. These mutant genes were used to reconstitute in vivo viral ribonucleoproteins (RNPs) whose biological activity was determined by (i) assay of viral RNA, cRNA, and mRNA accumulation in vivo, (ii) cap-dependent transcription in vitro, and (iii) cap snatching with purified recombinant RNPs. The results indicated that the W49A, F130A, and R142A mutations of PB2 reduced or abolished the capacity of mutant RNPs to synthesize RNA in vivo but did not substantially alter their ability to transcribe or carry out cap snatching in vitro. Some of the mutations (F130Y, R142A, and R142K) were rescued into infectious virus. While the F130Y mutant virus replicated faster than the wild type, mutant viruses R142A and R142K showed a delayed accumulation of cRNA and viral RNA during the infection cycle but normal kinetics of primary transcription, as determined by the accumulation of viral mRNA in cells infected in the presence of cycloheximide. These results indicate that the N-terminal region of PB2 plays a role in viral RNA replication.
Figures

Similar articles
-
The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication.PLoS One. 2008;3(12):e3904. doi: 10.1371/journal.pone.0003904. Epub 2008 Dec 10. PLoS One. 2008. PMID: 19066626 Free PMC article.
-
The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis.J Virol. 2000 Feb;74(3):1307-12. doi: 10.1128/jvi.74.3.1307-1312.2000. J Virol. 2000. PMID: 10627541 Free PMC article.
-
Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses.J Virol. 2003 May;77(10):6007-13. doi: 10.1128/jvi.77.10.6007-6013.2003. J Virol. 2003. PMID: 12719592 Free PMC article.
-
Structural insights into RNA synthesis by the influenza virus transcription-replication machine.Virus Res. 2017 Apr 15;234:103-117. doi: 10.1016/j.virusres.2017.01.013. Epub 2017 Jan 20. Virus Res. 2017. PMID: 28115197 Review.
-
Structure and Function of Influenza Virus Ribonucleoprotein.Subcell Biochem. 2018;88:95-128. doi: 10.1007/978-981-10-8456-0_5. Subcell Biochem. 2018. PMID: 29900494 Review.
Cited by
-
Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003 to 2005).J Clin Microbiol. 2006 Mar;44(3):1123-6. doi: 10.1128/JCM.44.3.1123-1126.2006. J Clin Microbiol. 2006. PMID: 16517910 Free PMC article.
-
The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication.PLoS One. 2008;3(12):e3904. doi: 10.1371/journal.pone.0003904. Epub 2008 Dec 10. PLoS One. 2008. PMID: 19066626 Free PMC article.
-
Involvement of influenza virus PA subunit in assembly of functional RNA polymerase complexes.J Virol. 2005 Jan;79(2):732-44. doi: 10.1128/JVI.79.2.732-744.2005. J Virol. 2005. PMID: 15613301 Free PMC article.
-
A big role for small RNAs in influenza virus replication.Proc Natl Acad Sci U S A. 2010 Jun 22;107(25):11153-4. doi: 10.1073/pnas.1006673107. Epub 2010 Jun 14. Proc Natl Acad Sci U S A. 2010. PMID: 20547839 Free PMC article. No abstract available.
-
Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein.J Virol. 2004 Apr;78(8):3880-8. doi: 10.1128/jvi.78.8.3880-3888.2004. J Virol. 2004. PMID: 15047804 Free PMC article.
References
-
- Asano, Y., and A. Ishihama. 1997. Identification of two nucleotide binding domains on the PB1 subunit of influenza virus RNA polymerase. J. Biochem. 122:627-634. - PubMed
-
- Asano, Y., K. Mizumoto, T. Maruyama, and A. Ishihama. 1995. Photoaffinity labeling of influenza virus RNA polymerase PB1 subunit with 8-azido GTP. J. Biochem. Tokyo 117:677-682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

