A long terminal repeat-containing retrotransposon of Schizosaccharomyces pombe expresses a Gag-like protein that assembles into virus-like particles which mediate reverse transcription
- PMID: 12692246
- PMCID: PMC153967
- DOI: 10.1128/jvi.77.9.5451-5463.2003
A long terminal repeat-containing retrotransposon of Schizosaccharomyces pombe expresses a Gag-like protein that assembles into virus-like particles which mediate reverse transcription
Abstract
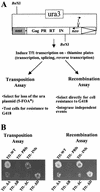
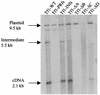
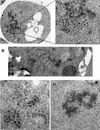
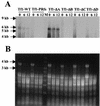
The Tf1 element of Schizosaccharomyces pombe is a long terminal repeat-containing retrotransposon that encodes functional protease, reverse transcriptase, and integrase proteins. Although these proteins are known to be necessary for protein processing, reverse transcription, and integration, respectively, the function of the protein thought to be Gag has not been determined. We present here the first electron microscopy of Tf1 particles. We tested whether the putative Gag of Tf1 was required for particle formation, packaging of RNA, and reverse transcription. We generated deletions of 10 amino acids in each of the four hydrophilic domains of the protein and found that all four mutations reduced transposition activity. The N-terminal deletion removed a nuclear localization signal and inhibited nuclear import of the transposon. The two mutations in the center of Gag destabilized the protein and resulted in no virus-like particles. The C-terminal deletion caused a defect in RNA packaging and, as a result, low levels of cDNA. The electron microscopy of cells expressing a truncated Tf1 showed that Gag alone was sufficient for the formation of virus-like particles. Taken together, these results indicate that Tf1 encodes a Gag protein that is a functional equivalent of the Gag proteins of retroviruses.
Figures



Similar articles
-
The long terminal repeat-containing retrotransposon Tf1 possesses amino acids in gag that regulate nuclear localization and particle formation.J Virol. 2005 Aug;79(15):9540-55. doi: 10.1128/JVI.79.15.9540-9555.2005. J Virol. 2005. PMID: 16014916 Free PMC article.
-
Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p.Mol Cell Biol. 2000 Oct;20(20):7798-812. doi: 10.1128/MCB.20.20.7798-7812.2000. Mol Cell Biol. 2000. PMID: 11003674 Free PMC article.
-
Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1.Mol Cell Biol. 1999 Aug;19(8):5768-84. doi: 10.1128/MCB.19.8.5768. Mol Cell Biol. 1999. PMID: 10409764 Free PMC article.
-
The Long Terminal Repeat Retrotransposons Tf1 and Tf2 of Schizosaccharomyces pombe.Microbiol Spectr. 2015 Aug;3(4):10.1128/microbiolspec.MDNA3-0040-2014. doi: 10.1128/microbiolspec.MDNA3-0040-2014. Microbiol Spectr. 2015. PMID: 26350316 Free PMC article. Review.
-
Domesticated DNA transposon proteins mediate retrotransposon control.Cell Res. 2008 Mar;18(3):331-3. doi: 10.1038/cr.2008.34. Cell Res. 2008. PMID: 18311163 Free PMC article. Review. No abstract available.
Cited by
-
The long terminal repeat-containing retrotransposon Tf1 possesses amino acids in gag that regulate nuclear localization and particle formation.J Virol. 2005 Aug;79(15):9540-55. doi: 10.1128/JVI.79.15.9540-9555.2005. J Virol. 2005. PMID: 16014916 Free PMC article.
-
Host factors that promote retrotransposon integration are similar in distantly related eukaryotes.PLoS Genet. 2017 Dec 12;13(12):e1006775. doi: 10.1371/journal.pgen.1006775. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29232693 Free PMC article.
-
The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr.Mol Biol Cell. 2005 Apr;16(4):1823-38. doi: 10.1091/mbc.e04-07-0583. Epub 2005 Jan 19. Mol Biol Cell. 2005. PMID: 15659641 Free PMC article.
-
LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex.Mol Cell Biol. 2007 Sep;27(18):6469-83. doi: 10.1128/MCB.00332-07. Epub 2007 Jun 11. Mol Cell Biol. 2007. PMID: 17562864 Free PMC article.
-
The yeast Ty1 retrotransposon requires components of the nuclear pore complex for transcription and genomic integration.Nucleic Acids Res. 2018 Apr 20;46(7):3552-3578. doi: 10.1093/nar/gky109. Nucleic Acids Res. 2018. PMID: 29514267 Free PMC article.
References
-
- Adams, S. E., J. Mellor, K. Gull, R. B. Sim, M. F. Tuite, S. M. Kingsman, and A. J. Kingsman. 1987. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian proteins. Cell 49:111-119. - PubMed
-
- Alin, K., and S. P. Goff. 1996. Mutational analysis of interactions between the Gag precursor proteins of murine leukemia viruses. Virology 216:418-424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

