Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments
- PMID: 12692259
- PMCID: PMC153940
- DOI: 10.1128/jvi.77.9.5540-5546.2003
Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments
Abstract
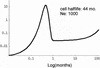
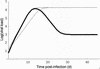
In vivo virologic compartments are cell types or tissues between which there is a restriction of virus flow, while virologic reservoirs are cell types or tissues in which there is a relative restriction of replication. The distinction between reservoirs and compartments is important because therapies that would be effective against a reservoir may not be effective against viruses produced by a given compartment, and vice versa. For example, the use of cytokines to "flush out" long-lived infected cells in patients on highly active antiretroviral therapy (T. W. Chun, D. Engel, M. M. Berrey, T. Shea, L. Corey, and A. S. Fauci, Proc. Natl. Acad. Sci. USA 95:8869-8873, 1998) may be successful for a latent reservoir but may not impact a compartment in which virus continues to replicate because of poor drug penetration. Here, we suggest phylogenetic criteria to illustrate, define, and differentiate between reservoirs and compartments. We then apply these criteria to the analysis of simulated and actual human immunodeficiency virus type 1 sequence data sets. We report that existing statistical methods work quite well at detecting viral compartments, and we learn from simulations that viral divergence from a calculated most recent common ancestor is a strong predictor of viral reservoirs.
Figures



Similar articles
-
Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure.J Virol. 2002 Sep;76(18):9481-92. doi: 10.1128/jvi.76.18.9481-9492.2002. J Virol. 2002. PMID: 12186930 Free PMC article.
-
Slow human immunodeficiency virus type 1 evolution in viral reservoirs in infants treated with effective antiretroviral therapy.AIDS Res Hum Retroviruses. 2007 Mar;23(3):381-90. doi: 10.1089/aid.2006.0175. AIDS Res Hum Retroviruses. 2007. PMID: 17411371
-
Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy.Annu Rev Immunol. 2000;18:665-708. doi: 10.1146/annurev.immunol.18.1.665. Annu Rev Immunol. 2000. PMID: 10837072 Review.
-
A novel assay allows genotyping of the latent reservoir for human immunodeficiency virus type 1 in the resting CD4+ T cells of viremic patients.J Virol. 2005 Apr;79(8):5185-202. doi: 10.1128/JVI.79.8.5185-5202.2005. J Virol. 2005. PMID: 15795302 Free PMC article.
-
The challenge of viral reservoirs in HIV-1 infection.Annu Rev Med. 2002;53:557-93. doi: 10.1146/annurev.med.53.082901.104024. Annu Rev Med. 2002. PMID: 11818490 Review.
Cited by
-
Redefining the viral reservoirs that prevent HIV-1 eradication.Immunity. 2012 Sep 21;37(3):377-88. doi: 10.1016/j.immuni.2012.08.010. Immunity. 2012. PMID: 22999944 Free PMC article. Review.
-
Human immunodeficiency virus (HIV) latency: the major hurdle in HIV eradication.Mol Med. 2012 Sep 25;18(1):1096-108. doi: 10.2119/molmed.2012.00194. Mol Med. 2012. PMID: 22692576 Free PMC article. Review.
-
HIV-1 Populations in Semen Arise through Multiple Mechanisms.PLoS Pathog. 2010 Aug 19;6(8):e1001053. doi: 10.1371/journal.ppat.1001053. PLoS Pathog. 2010. PMID: 20808902 Free PMC article.
-
Insights into the Impact of CD8+ Immune Modulation on Human Immunodeficiency Virus Evolutionary Dynamics in Distinct Anatomical Compartments by Using Simian Immunodeficiency Virus-Infected Macaque Models of AIDS Progression.J Virol. 2017 Nov 14;91(23):e01162-17. doi: 10.1128/JVI.01162-17. Print 2017 Dec 1. J Virol. 2017. PMID: 28931681 Free PMC article.
-
Restriction of HIV-1 genotypes in breast milk does not account for the population transmission genetic bottleneck that occurs following transmission.PLoS One. 2010 Apr 20;5(4):e10213. doi: 10.1371/journal.pone.0010213. PLoS One. 2010. PMID: 20422033 Free PMC article.
References
-
- Beerli, P., N. C. Grassly, M. K. Kuhner, D. Nickle, O. Pybus, M. Rain, A. Rambaut, A. G. Rodrigo, and Y. Wang. 2000. Population genetics of HIV: parameter estimation using genealogy-based methods, p. 217-258. In A. G. Rodrigo and G. H. Learn (ed.), Computational and evolutionary analyses of HIV sequences. Kluwer Academic Publishers, Boston, Mass.
-
- Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. - PubMed
-
- Bourinbaiar, A. S. 1994. The ratio of defective HIV-1 particles to replication-competent infectious virions. Acta Virol. 38:59-61. - PubMed
-
- Chun, T. W., R. T. Davey, Jr., M. Ostrowski, J. S. Justement, D. Engel, J. I. Mullins, and A. S. Fauci. 2000. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6:757-761. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

