Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis
- PMID: 12694625
- PMCID: PMC1992516
- DOI: 10.1046/j.1365-2958.2003.03474.x
Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis
Abstract
Unlike many pathogens that are overtly harmful to their hosts, Mycobacterium tuberculosis can persist for years within humans in a clinically latent state. Latency is often linked to hypoxic conditions within the host. Among M. tuberculosis genes induced by hypoxia is a putative transcription factor, Rv3133c/DosR. We performed targeted disruption of this locus followed by transcriptome analysis of wild-type and mutant bacilli. Nearly all the genes powerfully regulated by hypoxia require Rv3133c/DosR for their induction. Computer analysis identified a consensus motif, a variant of which is located upstream of nearly all M. tuberculosis genes rapidly induced by hypoxia. Further, Rv3133c/DosR binds to the two copies of this motif upstream of the hypoxic response gene alpha-crystallin. Mutations within the binding sites abolish both Rv3133c/DosR binding as well as hypoxic induction of a downstream reporter gene. Also, mutation experiments with Rv3133c/DosR confirmed sequence-based predictions that the C-terminus is responsible for DNA binding and that the aspartate at position 54 is essential for function. Together, these results demonstrate that Rv3133c/DosR is a transcription factor of the two-component response regulator class, and that it is the primary mediator of a hypoxic signal within M. tuberculosis.
Figures

SDS-PAGE analysis of DosR expression in E. coli. The dosR coding sequence was cloned into a pET expression vector, and expression was induced by IPTG. Lanes (1–3) full length: 1, uninduced; 2, induced for 2 h; 3, induced for 5 h; lanes (4–5) amino acids 1–134: 4, uninduced; 5, induced for 5 h; lanes (6–7) amino acids 144–217: 6, uninduced; 7, induced for 5 h. Arrows indicate the position at which DosR is expected to migrate.
Electrophoretic mobility shift assay (EMSA) of E. coli extracts with full-length, N-terminal and C-terminal portions of DosR and radiolabelled acr promoter DNA. Lanes: 1, no extract; 2, amino acids 1–217 (full length); 3, amino acids 1–134; 4, amino acids 144–217.
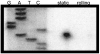

EMSA of E. coli extracts and radiolabelled overlapping regions of acr promoter DNA. Labelled probe A (−149 to −96); B (−111 to −79); C (−89 to −49); D (−66 to −34). For each fragment, DNA was mixed with (1) no extract, (2) uninduced extract and (3) induced extract.
Schematic diagram of acr promoter (solid black line) with regions analysed (dashed lines) relative to hypoxia motif sites (black boxes).

20-mer hypoxia motifs in the native acr promoter. Positions are relative to the transcription start site. Underlined bases were mutated as follows: ‘ccaa’→‘ggtt’ and ‘tcgg’→‘agcc’.
EMSA with radiolabelled probes and specific unlabelled competitor DNAs. Lanes 1–6, labelled probe (−111 to −79); lanes 7–12, labelled probe (−66 to −34). Lanes 1 and 7, no extract control. Lanes 2 and 8, no competitor control. Lanes 3 and 11, unlabelled competitor (−111 to −79). Lanes 4 and 12, unlabelled competitor (mutant −111 to −79). Lanes 5 and 9, unlabelled competitor (−66 to −34). Lanes 6 and 10, unlabelled competitor (mutant −66 to −34).
Hypoxic induction of acr promoter with mutations in the hypoxia motifs. Activity of acr promoter was measured by luciferase reporter gene assay. Indicated mutations in distal and proximal motifs were made as described above. Shown are representative data from one of two experiments, each of which was performed in duplicate.

EMSA of mutant DosR (Asp-54Glu) with labelled acr promoter DNA. Lanes: 1, no extract; 2, induced extract of DosR (Asp-54Glu); 3, induced extract of DosR (WT).
Hypoxic induction of acr by mutant DosR (Asp-54Glu). Acr expression measured by luciferase reporter gene in BCG and in BCG with DosR (Asp-54Glu) replacing DosR (WT). Shown are representative data from one of two experiments, each of which was performed in duplicate.
References
-
- Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. - PubMed
-
- Bloom BR, Small PM. The evolving relation between humans and Mycobacterium tuberculosis. N Engl J Med. 1998;338:677–678. - PubMed
-
- Canetti G. The Tubercle Bacillus in the Pulmonary Lesion of Man. Springer Publishing; New York: 1955. Growth of the tubercle bacillus in the tuberculosis lesion; pp. 111–126.
-
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

