2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency is caused by mutations in the HADH2 gene
- PMID: 12696021
- PMCID: PMC1180283
- DOI: 10.1086/375116
2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency is caused by mutations in the HADH2 gene
Abstract
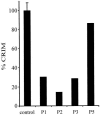
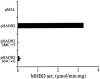
2-methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency is a novel inborn error of isoleucine degradation. In this article, we report the elucidation of the molecular basis of MHBD deficiency. To this end, we purified the enzyme from bovine liver. MALDI-TOF mass spectrometry analysis revealed that the purified protein was identical to bovine 3-hydroxyacyl-CoA dehydrogenase type II. The human homolog of this bovine enzyme is a short-chain 3-hydroxyacyl-CoA dehydrogenase, also known as the "endoplasmic reticulum-associated amyloid-beta binding protein" (ERAB). This led to the identification of the X-chromosomal gene involved, which previously had been denoted "HADH2." Sequence analysis of the HADH2 gene from patients with MHBD deficiency revealed the presence of two missense mutations (R130C and L122V). Heterologous expression of the mutant cDNAs in Escherichia coli showed that both mutations almost completely abolish enzyme activity. This confirms that MHBD deficiency is caused by mutations in the HADH2 gene.
Figures



References
Electronic-Database Information
-
- GenBank and National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/ (for ERAB [accession numbers U96132 and AF037438] and HADH2 [NM_004493])
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for 2-methylacetoacetyl-CoA thiolase deficiency, 2-methylbutyryl-CoA dehydrogenase deficiency, 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency, and Alzheimer disease)
-
- Swiss-Prot Protein Knowledgebase, http://www.ebi.ac.uk/swissprot/ (for a protein sequence database)
References
-
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 - PubMed
-
- Ensenauer R, Niederhoff H, Ruiter JP, Wanders RJ, Schwab KO, Brandis M, Lehnert W (2002) Clinical variability in 3-hydroxy-2-methylbutyryl-CoA dehydrogenase deficiency. Ann Neurol 51:656–659 - PubMed
-
- Frackowiak J, Mazur-Kolecka B, Kaczmarski W, Dickson D (2001) Deposition of Alzheimer’s vascular amyloid-β is associated with decreased expression of brain L-3-hydroxyacyl-coenzyme A dehydrogenase (ERAB). Brain Res 907:44–53 - PubMed
-
- Furuta S, Kobayashi A, Miyazawa S, Hashimoto T (1997) Cloning and expression of cDNA for a newly identified isozyme of bovine liver 3-hydroxyacyl-CoA dehydrogenase and its import into mitochondria. Biochim Biophys Acta 1350:317–324 - PubMed
-
- Ghaedi K, Itagaki A, Toyama R, Tamura S, Matsumura T, Kawai A, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y (1999) Newly identified Chinese hamster ovary cell mutants defective in peroxisome assembly represent complementation group A of human peroxisome biogenesis disorders and one novel group in mammals. Exp Cell Res 248:482–488 - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

