IfkA, a presumptive eIF2 alpha kinase of Dictyostelium, is required for proper timing of aggregation and regulation of mound size
- PMID: 12697064
- PMCID: PMC154100
- DOI: 10.1186/1471-213x-3-3
IfkA, a presumptive eIF2 alpha kinase of Dictyostelium, is required for proper timing of aggregation and regulation of mound size
Abstract
Background: The transition from growth to development in Dictyostelium is initiated by amino acid starvation of growing amobae. In other eukaryotes, a key sensor of amino acid starvation and mediator of the resulting physiological responses is the GCN2 protein, an eIF2alpha kinase. GCN2 downregulates the initiation of translation of bulk mRNA and enhances translation of specific mRNAs by phosphorylating the translation initiation factor eIF2alpha. Two eIF2alpha kinases were identified in Dictyostelium and studied herein.
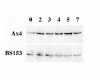
Results: Neither of the eIF2alpha kinases appeared to be involved in sensing amino acid starvation to initiate development. However, one of the kinases, IfkA, was shown to phosphorylate eIF2alpha from 1 to 7 hours after the onset of development, resulting in a shift from polysomes to free ribosomes for bulk mRNA. In the absence of the eIF2alpha phosphorylation, ifkA null cells aggregated earlier than normal and formed mounds and ultimately fruiting bodies that were larger than normal. The early aggregation phenotype in ifkA null cells reflected an apparent, earlier than normal establishment of the cAMP pulsing system. The large mound phenotype resulted from a reduced extracellular level of Countin, a component of the counting factor that regulates mound size. In wild type cells, phosphorylation of eIF2alpha by IfkA resulted in a specific stabilization and enhanced translational efficiency of countin mRNA even though reduced translation resulted for bulk mRNA.
Conclusions: IfkA is an eIF2alpha kinase of Dictyostelium that normally phosphorylates eIF2alpha from 1 to 7 hours after the onset of development, or during the preaggregation phase. This results in an overall reduction in the initiation of protein synthesis during this time frame and a concomitant reduction in the number of ribosomes associated with most mRNAs. For some mRNAs, however, initiation of protein synthesis is enhanced or stabilized under the conditions of increased eIF2alpha phosphorylation. This includes countin mRNA.
Figures








References
-
- Raper KB. The Dictyostelids. Princeton University Press, Princeton, NJ. 1984.
-
- Gomer RH, Yuen IS, Firtel RA. A secreted 80*10(3)Mr protein mediates sensing of cell density and the onset of development in Dictyostelium. Develop. 1991;112:269. - PubMed
-
- Yuen IS, Taphouse C, Halfant KA, Gomer RH. Regulation and processing of a secreted protein that mediates sensing of cell density in Dictyostelium. Develop. 1991;113:1375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

