Comparative genomic tools and databases: providing insights into the human genome
- PMID: 12697725
- PMCID: PMC152942
- DOI: 10.1172/JCI17842
Comparative genomic tools and databases: providing insights into the human genome
Figures
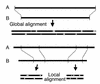



Similar articles
-
Paircomp, FamilyRelationsII and Cartwheel: tools for interspecific sequence comparison.BMC Bioinformatics. 2005 Mar 24;6:70. doi: 10.1186/1471-2105-6-70. BMC Bioinformatics. 2005. PMID: 15790396 Free PMC article.
-
From phenotyping to genotyping - bioinformatics for the busy clinician.Eur J Med Genet. 2019 Aug;62(8):103689. doi: 10.1016/j.ejmg.2019.103689. Epub 2019 Jun 18. Eur J Med Genet. 2019. PMID: 31226441 Review.
-
A tiered approach to comparative genomics.Brief Funct Genomic Proteomic. 2005 Jul;4(2):178-85. doi: 10.1093/bfgp/4.2.178. Brief Funct Genomic Proteomic. 2005. PMID: 16102272 Review.
-
Genome resources and comparative analysis tools for cardiovascular research.Methods Mol Med. 2006;128:101-23. doi: 10.1007/978-1-59745-159-8_8. Methods Mol Med. 2006. PMID: 17071992
-
Ensembl 2002: accommodating comparative genomics.Nucleic Acids Res. 2003 Jan 1;31(1):38-42. doi: 10.1093/nar/gkg083. Nucleic Acids Res. 2003. PMID: 12519943 Free PMC article.
Cited by
-
Identifying transcriptional targets.Genome Biol. 2004;5(3):210. doi: 10.1186/gb-2004-5-3-210. Epub 2004 Feb 27. Genome Biol. 2004. PMID: 15005803 Free PMC article. Review.
-
Comparative genomic analysis as a tool for biological discovery.J Physiol. 2004 Jan 1;554(Pt 1):31-9. doi: 10.1113/jphysiol.2003.050948. J Physiol. 2004. PMID: 14678488 Free PMC article. Review.
-
Bridging cancer biology with the clinic: relative expression of a GRHL2-mediated gene-set pair predicts breast cancer metastasis.PLoS One. 2013;8(2):e56195. doi: 10.1371/journal.pone.0056195. Epub 2013 Feb 18. PLoS One. 2013. PMID: 23441166 Free PMC article.
-
Construction of predictive promoter models on the example of antibacterial response of human epithelial cells.Theor Biol Med Model. 2005 Jan 12;2:2. doi: 10.1186/1742-4682-2-2. Theor Biol Med Model. 2005. PMID: 15647113 Free PMC article.
-
Marine organism cell biology and regulatory sequence discoveryin comparative functional genomics.Cytotechnology. 2004 Oct;46(2-3):123-37. doi: 10.1007/s10616-005-1719-5. Epub 2005 Nov 30. Cytotechnology. 2004. PMID: 19003267 Free PMC article.
References
-
- Mayor C, et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. - PubMed
-
- Grehan S, Allan C, Tse E, Walker D, Taylor JM. Expression of the apolipoprotein E gene in the skin is controlled by a unique downstream enhancer. J. Invest. Dermatol. 2001;116:77–84. - PubMed

