Simulating disorder-order transitions in molecular recognition of unstructured proteins: where folding meets binding
- PMID: 12697905
- PMCID: PMC154313
- DOI: 10.1073/pnas.0531373100
Simulating disorder-order transitions in molecular recognition of unstructured proteins: where folding meets binding
Abstract
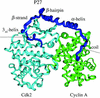
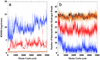
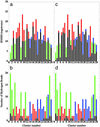
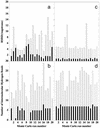
A microscopic study of functional disorder-order folding transitions coupled to binding is performed for the p27 protein, which derives a kinetic advantage from the intrinsically disordered unbound form on binding with the phosphorylated cyclin A-cyclin-dependent kinase 2 (Cdk2) complex. Hierarchy of structural loss during p27 coupled unfolding and unbinding is simulated by using high-temperature Monte Carlo simulations initiated from the crystal structure of the tertiary complex. Subsequent determination of the transition-state ensemble and the proposed atomic picture of the folding mechanism coupled to binding provide a microscopic rationale that reconciles the initiation recruitment of p27 at the cyclin A docking site with the kinetic benefit for a disordered alpha-helix in the unbound form of p27. The emerging structural polarization in the ensemble of unfolding/unbinding trajectories and in the computationally determined transition-state ensemble is not determined by the intrinsic folding preferences of p27 but rather is attributed to the topological requirements of the native intermolecular interface to order beta-hairpin and beta-strand of p27 that could be critical for nucleating rapid folding transition coupled to binding. In agreement with the experimental data, the disorder-order folding transition for p27 is largely determined by the functional requirement to form a specific intermolecular interface that ultimately dictates the folding mechanism and overwhelms any local folding preferences for creating a stable alpha-helix in the p27 structure before overcoming the major free energy barrier.
Figures

References
-
- Wright P E, Dyson H J. J Mol Biol. 1999;293:321–331. - PubMed
-
- Dunker A K, Lawson J D, Brown C J, Williams R M, Romero P, Oh J S, Oldfield C J, Campen A M, Obradovic Z. J Mol Graphics Model. 2001;19:26–59. - PubMed
-
- Dyson H J, Wright P E. Curr Opin Struct Biol. 2002;12:54–60. - PubMed
-
- Spolar R S, Record M T. Science. 1994;263:777–784. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

