Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea
- PMID: 12700255
- PMCID: PMC154410
- DOI: 10.1128/JB.185.9.2759-2773.2003
Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea
Erratum in
- J Bacteriol. 2003 Nov;185(21):6496
Abstract
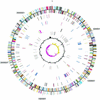
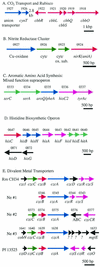
Nitrosomonas europaea (ATCC 19718) is a gram-negative obligate chemolithoautotroph that can derive all its energy and reductant for growth from the oxidation of ammonia to nitrite. Nitrosomonas europaea participates in the biogeochemical N cycle in the process of nitrification. Its genome consists of a single circular chromosome of 2,812,094 bp. The GC skew analysis indicates that the genome is divided into two unequal replichores. Genes are distributed evenly around the genome, with approximately 47% transcribed from one strand and approximately 53% transcribed from the complementary strand. A total of 2,460 protein-encoding genes emerged from the modeling effort, averaging 1,011 bp in length, with intergenic regions averaging 117 bp. Genes necessary for the catabolism of ammonia, energy and reductant generation, biosynthesis, and CO(2) and NH(3) assimilation were identified. In contrast, genes for catabolism of organic compounds are limited. Genes encoding transporters for inorganic ions were plentiful, whereas genes encoding transporters for organic molecules were scant. Complex repetitive elements constitute ca. 5% of the genome. Among these are 85 predicted insertion sequence elements in eight different families. The strategy of N. europaea to accumulate Fe from the environment involves several classes of Fe receptors with more than 20 genes devoted to these receptors. However, genes for the synthesis of only one siderophore, citrate, were identified in the genome. This genome has provided new insights into the growth and metabolism of ammonia-oxidizing bacteria.
Figures
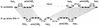


Comment in
-
Lessons from the genome of a lithoautotroph: making biomass from almost nothing.J Bacteriol. 2003 May;185(9):2690-1. doi: 10.1128/JB.185.9.2690-2691.2003. J Bacteriol. 2003. PMID: 12700247 Free PMC article. No abstract available.
Similar articles
-
Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation.Environ Microbiol. 2007 Dec;9(12):2993-3007. doi: 10.1111/j.1462-2920.2007.01409.x. Environ Microbiol. 2007. PMID: 17991028
-
Genome sequence of Nitrosomonas sp. strain AL212, an ammonia-oxidizing bacterium sensitive to high levels of ammonia.J Bacteriol. 2011 Sep;193(18):5047-8. doi: 10.1128/JB.05521-11. J Bacteriol. 2011. PMID: 21868805 Free PMC article.
-
Steady-State Growth under Inorganic Carbon Limitation Conditions Increases Energy Consumption for Maintenance and Enhances Nitrous Oxide Production in Nitrosomonas europaea.Appl Environ Microbiol. 2016 May 16;82(11):3310-3318. doi: 10.1128/AEM.00294-16. Print 2016 Jun 1. Appl Environ Microbiol. 2016. PMID: 27016565 Free PMC article.
-
Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea.Arch Microbiol. 2002 Oct;178(4):250-5. doi: 10.1007/s00203-002-0452-0. Epub 2002 Jun 27. Arch Microbiol. 2002. PMID: 12209257 Review.
-
The impact of genome analyses on our understanding of ammonia-oxidizing bacteria.Annu Rev Microbiol. 2007;61:503-28. doi: 10.1146/annurev.micro.61.080706.093449. Annu Rev Microbiol. 2007. PMID: 17506671 Review.
Cited by
-
Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility.Proc Natl Acad Sci U S A. 2006 Oct 17;103(42):15280-7. doi: 10.1073/pnas.0606924103. Epub 2006 Oct 9. Proc Natl Acad Sci U S A. 2006. PMID: 17030797 Free PMC article.
-
Genome analysis of Minibacterium massiliensis highlights the convergent evolution of water-living bacteria.PLoS Genet. 2007 Aug;3(8):e138. doi: 10.1371/journal.pgen.0030138. Epub 2007 Jul 5. PLoS Genet. 2007. PMID: 17722982 Free PMC article.
-
Identification of acyl-homoserine lactone signal molecules produced by Nitrosomonas europaea strain Schmidt.Appl Environ Microbiol. 2005 Aug;71(8):4906-9. doi: 10.1128/AEM.71.8.4906-4909.2005. Appl Environ Microbiol. 2005. PMID: 16085894 Free PMC article.
-
Diversity, abundance, and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China.Appl Environ Microbiol. 2010 Jul;76(14):4691-702. doi: 10.1128/AEM.02563-09. Epub 2010 May 28. Appl Environ Microbiol. 2010. PMID: 20511433 Free PMC article.
-
Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach.Ecotoxicology. 2018 Nov;27(9):1281-1291. doi: 10.1007/s10646-018-1981-x. Epub 2018 Sep 21. Ecotoxicology. 2018. PMID: 30242595
References
-
- Abeliovich, A., and A. Vonsahk. 1992. Anaerobic metabolism of Nitrosomonas europaea. Arch. Microbiol. 158:267-270.
-
- Arciero, D., and A. B. Hooper. 1994. A di-heme cytochrome c peroxidase from Nitrosomonas europaea catalytically active in both the oxidized and half-reduced states. J. Biol. Chem. 269:11878-11886. - PubMed
-
- Badger, J. H., and G. J. Olsen. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512-524. - PubMed
-
- Beaumont, H. J., N. G. Hommes, L. A. Sayavedra-Soto, D. J. Arp, D. M. Arciero, A. B. Hooper, H. V. Westerhoff, and R. J. van Spanning. 2002. Nitrite reductase of Nitrosomonas europaea is not essential for production of gaseous nitrogen oxides and confers tolerance to nitrite. J. Bacteriol. 184:2557-2560. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

