Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol
- PMID: 12700259
- PMCID: PMC154405
- DOI: 10.1128/JB.185.9.2802-2810.2003
Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol
Abstract
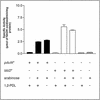
The studies reported here identify propionyl coenzyme A (propionyl-CoA) as the common intermediate in the 1,2-propanediol and propionate catabolic pathways of Salmonella enterica serovar Typhimurium LT2. Growth on 1,2-propanediol as a carbon and energy source led to the formation and excretion of propionate, whose activation to propionyl-CoA relied on the activities of the propionate kinase (PduW)/phosphotransacetylase (Pta) enzyme system and the CobB sirtuin-controlled acetyl-CoA and propionyl-CoA (Acs, PrpE) synthetases. The different affinities of these systems for propionate ensure sufficient synthesis of propionyl-CoA to support wild-type growth of S. enterica under low or high concentrations of propionate in the environment. These redundant systems of propionyl-CoA synthesis are needed because the prpE gene encoding the propionyl-CoA synthetase enzyme is part of the prpBCDE operon under the control of the PrpR regulatory protein, which needs 2-methylcitrate as a coactivator. Because the synthesis of 2-methylcitrate by PrpC (i.e., the 2-methylcitrate synthase enzyme) requires propionyl-CoA as a substrate, the level of propionyl-CoA needs to be raised by the Acs or PduW-Pta system before 2-methylcitrate can be synthesized and prpBCDE transcription can be activated.
Figures



References
-
- Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. - PMC - PubMed
-
- Brämer, C. O., L. F. Silva, J. G. C. Gomez, H. Priefert, and A. Steinbüchel. 2002. Identification of the 2-methylcitrate pathway involved in the catabolism of propionate in the polyhydroxyalkanoate-producing strain Burkholderia sacchari IPT101T and analysis of a mutant accumulating a copolyester with higher 3-hydroxyvalerate content. Appl. Environ. Microbiol. 68:271-279. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

