Analysis and functional classification of transcripts from the nematode Meloidogyne incognita
- PMID: 12702207
- PMCID: PMC154577
- DOI: 10.1186/gb-2003-4-4-r26
Analysis and functional classification of transcripts from the nematode Meloidogyne incognita
Abstract
Background: Plant parasitic nematodes are major pathogens of most crops. Molecular characterization of these species as well as the development of new techniques for control can benefit from genomic approaches. As an entrée to characterizing plant parasitic nematode genomes, we analyzed 5,700 expressed sequence tags (ESTs) from second-stage larvae (L2) of the root-knot nematode Meloidogyne incognita.
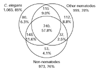
Results: From these, 1,625 EST clusters were formed and classified by function using the Gene Ontology (GO) hierarchy and the Kyoto KEGG database. L2 larvae, which represent the infective stage of the life cycle before plant invasion, express a diverse array of ligand-binding proteins and abundant cytoskeletal proteins. L2 are structurally similar to Caenorhabditis elegans dauer larva and the presence of transcripts encoding glyoxylate pathway enzymes in the M. incognita clusters suggests that root-knot nematode larvae metabolize lipid stores while in search of a host. Homology to other species was observed in 79% of translated cluster sequences, with the C. elegans genome providing more information than any other source. In addition to identifying putative nematode-specific and Tylenchida-specific genes, sequencing revealed previously uncharacterized horizontal gene transfer candidates in Meloidogyne with high identity to rhizobacterial genes including homologs of nodL acetyltransferase and novel cellulases.
Conclusions: With sequencing from plant parasitic nematodes accelerating, the approaches to transcript characterization described here can be applied to more extensive datasets and also provide a foundation for more complex genome analyses.
Figures


Similar articles
-
A survey of SL1-spliced transcripts from the root-lesion nematode Pratylenchus penetrans.Mol Genet Genomics. 2004 Sep;272(2):138-48. doi: 10.1007/s00438-004-1054-0. Epub 2004 Aug 28. Mol Genet Genomics. 2004. PMID: 15338281
-
Genomes of parasitic nematodes (Meloidogyne hapla, Meloidogyne incognita, Ascaris suum and Brugia malayi) have a reduced complement of small RNA interference pathway genes: knockdown can reduce host infectivity of M. incognita.Funct Integr Genomics. 2016 Jul;16(4):441-57. doi: 10.1007/s10142-016-0495-y. Epub 2016 Apr 28. Funct Integr Genomics. 2016. PMID: 27126863
-
Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita.Nat Biotechnol. 2008 Aug;26(8):909-15. doi: 10.1038/nbt.1482. Epub 2008 Jul 27. Nat Biotechnol. 2008. PMID: 18660804
-
The genomes of root-knot nematodes.Annu Rev Phytopathol. 2009;47:333-51. doi: 10.1146/annurev-phyto-080508-081839. Annu Rev Phytopathol. 2009. PMID: 19400640 Review.
-
Interactions between bacteria and plant-parasitic nematodes: now and then.Int J Parasitol. 2003 Sep 30;33(11):1269-76. doi: 10.1016/s0020-7519(03)00160-7. Int J Parasitol. 2003. PMID: 13678641 Review.
Cited by
-
Effector gene vap1 based DGGE fingerprinting to assess variation within and among Heterodera schachtii populations.J Nematol. 2018;50(4):517-528. doi: 10.21307/jofnem-2018-055. J Nematol. 2018. PMID: 31094153 Free PMC article.
-
Characterization of mRNA Expression and Endogenous RNA Profiles in Bladder Cancer Based on The Cancer Genome Atlas (TCGA) Database.Med Sci Monit. 2019 Apr 25;25:3041-3060. doi: 10.12659/MSM.915487. Med Sci Monit. 2019. PMID: 31020952 Free PMC article.
-
Sequence mining and transcript profiling to explore cyst nematode parasitism.BMC Genomics. 2009 Jan 30;10:58. doi: 10.1186/1471-2164-10-58. BMC Genomics. 2009. PMID: 19183474 Free PMC article.
-
Genome Expression Dynamics Reveal the Parasitism Regulatory Landscape of the Root-Knot Nematode Meloidogyne incognita and a Promoter Motif Associated with Effector Genes.Genes (Basel). 2021 May 18;12(5):771. doi: 10.3390/genes12050771. Genes (Basel). 2021. PMID: 34070210 Free PMC article.
-
Automated methods of predicting the function of biological sequences using GO and BLAST.BMC Bioinformatics. 2005 Nov 15;6:272. doi: 10.1186/1471-2105-6-272. BMC Bioinformatics. 2005. PMID: 16288652 Free PMC article.
References
-
- Agrios GN. Plant diseases caused by nematodes. In: Agrios GN, editor. In Plant Pathology. New York: Academic Press; 1997. pp. 565–597.
-
- Sasser JN, Freckman DW. A world perspective on nematology: the role of the society. In: Veech JA, Dickson DW, editor. In Vistas on Nematology. Hyattsville: Society of Nematology; 1987. pp. 7–14.
-
- Taylor AL, Sasser JN. Biology, Identification and Control of Root-knot Nematodes (Meloidogyne species) Raleigh, NC: United States Agency for International Development; 1978.
-
- Wyss U, Grundler FMW, Munch A. The parasitic behaviour of second-stage juveniles of Meloidogyne incognita in roots of Arabidopsis thaliana. Nematologica. 1992;38:98–111.
-
- Smant G, Stokkermans JP, Yan Y, de Boer JM, Baum TJ, Wang X, Hussey RS, Gommers FJ, Henrissat B, Davis EL, et al. Endogenous cellulases in animals: isolation of beta-1, 4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc Natl Acad Sci USA. 1998;95:4906–4911. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

