A quantitative analysis of intron effects on mammalian gene expression
- PMID: 12702819
- PMCID: PMC1370426
- DOI: 10.1261/rna.5250403
A quantitative analysis of intron effects on mammalian gene expression
Abstract
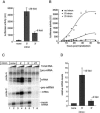
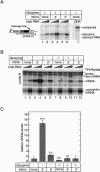
In higher eukaryotes, intron-containing and intronless versions of otherwise identical genes can exhibit dramatically different expression profiles. Introns and the act of their removal by the spliceosome can affect gene expression at many different levels, including transcription, polyadenylation, mRNA export, translational efficiency, and the rate of mRNA decay. However, the extent to which each of these steps contributes to the overall effect of any one intron on gene expression has not been rigorously tested. Here we report construction and initial characterization of a luciferase-based reporter system for monitoring the effects of individual introns and their position within the gene on protein expression in mammalian cells. Quantitative analysis of constructs containing human TPI intron 6 at two different positions within the Renilla luciferase open reading frame revealed that this intron acts primarily to enhance mRNA accumulation. Spliced mRNAs also exhibited higher translational yields than did intronless transcripts. However, nucleocytoplasmic mRNA distribution and mRNA stability were largely unaffected. These findings were extended to two other introns in a TCR-beta minigene.
Figures

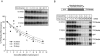

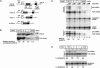
References
-
- Andrulis, E.D., Werner, J., Nazarian, A., Erdjument-Bromage, H., Tempst, P., and Lis, J.T. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837–841. - PubMed
-
- Bousquet-Antonelli, C., Presutti, C., and Tollervey, D. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775. - PubMed
-
- Bouvet, P. and Wolffe, A.P. 1994. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 77: 931–941. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
