Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells
- PMID: 12702820
- PMCID: PMC1370427
- DOI: 10.1261/rna.5260303
Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells
Abstract
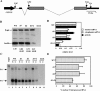
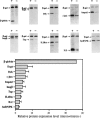
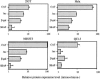
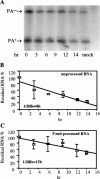
We have examined how splicing affects the expression of a range of human and nonhuman genes in vertebrate cells. Although our data demonstrate that splicing invariably enhances the level of gene expression, this positive effect is generally moderate. However, in the case of the human beta-globin gene, splicing is essential for significant protein expression. In the absence of introns, 3' end processing is inefficient, and this appears to be causally linked to a significant decrease in the level of both nuclear and cytoplasmic 3' end-processed RNA. In contrast, splicing appears to only modestly enhance nuclear mRNA export. Consistent with this observation, intronless beta-globin gene expression was only partially rescued by the insertion of retroviral nuclear mRNA export elements. Surprisingly, in the case of the highly intron dependent beta-globin gene, the mRNA that did reach the cytoplasm was also only inefficiently translated if it derived from an intronless expression plasmid. Together, these data argue that splicing can increase gene expression by enhancing mRNA 3' end processing, and hence, mRNA production. Moreover, in the case of the highly intron-dependent beta-globin gene, splicing also significantly enhanced the translational utilization of cytoplasmic beta-globin mRNAs.
Figures

References
-
- Chang, D.D. and Sharp, P.A. 1989. Regulation by HIV Rev depends upon recognition of splice sites. Cell 59: 789–795. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
