Complete genome sequence of the Q-fever pathogen Coxiella burnetii
- PMID: 12704232
- PMCID: PMC154366
- DOI: 10.1073/pnas.0931379100
Complete genome sequence of the Q-fever pathogen Coxiella burnetii
Abstract
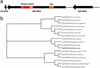
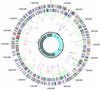
The 1,995,275-bp genome of Coxiella burnetii, Nine Mile phase I RSA493, a highly virulent zoonotic pathogen and category B bioterrorism agent, was sequenced by the random shotgun method. This bacterium is an obligate intracellular acidophile that is highly adapted for life within the eukaryotic phagolysosome. Genome analysis revealed many genes with potential roles in adhesion, invasion, intracellular trafficking, host-cell modulation, and detoxification. A previously uncharacterized 13-member family of ankyrin repeat-containing proteins is implicated in the pathogenesis of this organism. Although the lifestyle and parasitic strategies of C. burnetii resemble that of Rickettsiae and Chlamydiae, their genome architectures differ considerably in terms of presence of mobile elements, extent of genome reduction, metabolic capabilities, and transporter profiles. The presence of 83 pseudogenes displays an ongoing process of gene degradation. Unlike other obligate intracellular bacteria, 32 insertion sequences are found dispersed in the chromosome, indicating some plasticity in the C. burnetii genome. These analyses suggest that the obligate intracellular lifestyle of C. burnetii may be a relatively recent innovation.
Figures

References
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

