Dispersal of NK homeobox gene clusters in amphioxus and humans
- PMID: 12704239
- PMCID: PMC154338
- DOI: 10.1073/pnas.0836141100
Dispersal of NK homeobox gene clusters in amphioxus and humans
Abstract
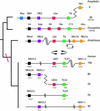
The Drosophila melanogaster genome has six physically clustered NK-related homeobox genes in just 180 kb. Here we show that the NK homeobox gene cluster was an ancient feature of bilaterian animal genomes, but has been secondarily split in chordate ancestry. The NK homeobox gene clusters of amphioxus and vertebrates are each split and dispersed at two equivalent intergenic positions. From the ancestral NK gene cluster, only the Tlx-Lbx and NK3-NK4 linkages have been retained in chordates. This evolutionary pattern is in marked contrast to the Hox and ParaHox gene clusters, which are compact in amphioxus and vertebrates, but have been disrupted in Drosophila.
Figures



References
-
- Jagla K, Jagla T, Heitzler P, Dretzen G, Bellard F, Bellard T. Development (Cambridge, UK) 1997;124:91–100. - PubMed
-
- Jagla K, Bellard M, Frasch M. BioEssays. 2001;23:125–133. - PubMed
-
- Galliot B, de Vargas C, Miller D. Dev Genes Evol. 1999;209:186–197. - PubMed
-
- Pollard S L, Holland P W H. Curr Biol. 2000;10:1059–1062. - PubMed
-
- Harvey R P. Dev Biol. 1996;178:203–216. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

