Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3
- PMID: 12706667
- PMCID: PMC7173220
- DOI: 10.1016/s0264-410x(03)00027-6
Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3
Abstract
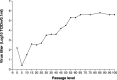
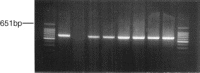
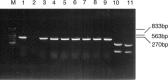
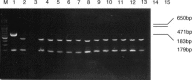
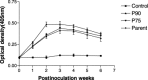
A porcine epidemic diarrhea virus (PEDV) designated DR13 was isolated in Vero cells and serially passaged by level 100. The virus was titrated at regular intervals of the passage level. Open reading frame (ORF) 3 sequences of the virus at passage levels 20, 40, 60, 80, and 100 were aligned and compared using a computer software program. Suitability of the restriction fragment length polymorphism (RFLP) analysis for differentiating the virus from other Korean field strains was investigated. The DR13 field isolate was successively adapted in Vero cells as observed through polymerase chain reaction (PCR) and titration of the virus. RFLP analysis identified change in cleavage sites of HindIII and Xho II from passage levels 75 and 90, respectively; these RFLP patterns of ORF 3 differentiated the Vero cell-adapted virus from its parent strain, DR13, and 12 other strains of PEDV studied. The cell adapted DR13 was tested for its pathogenicity and immunogenicity in piglets and pregnant sows. The results indicated that cell adapted DR13 revealed reduced pathogenicity and induced protective immune response in pigs. Differentiation between highly Vero cell-adapted virus and wild-type virus could be the marker of adaptation to cell culture and a valuable tool for epidemiologic studies of PEDV infections. The results of this study supported that the cell attenuated virus could be applied as a marker vaccine candidate against PEDV infection.
Figures




References
-
- Bernasconi C, Guscetti F, Utiger A, Van Reeth K, Ackermann M, Pospischil A. Experimental infection of genotobiotic piglets with a cell culture adapted porcine epidemic diarrhea virus: clinical, histopathological and immunohistochemical findings. In: Proceedings of the Third Congress of ESVV, Interlaken, Switzerland, 1995. p. 542–6.
-
- Bridgen A., Duarte M., Tobler K., Laude H., Ackermann M. Sequence determination of the nucleocapsid protein gene of the porcine epidemic diarrhea virus confirms that this virus is a coronavirus related to human coronavirus 229E and porcine transmissible gastroenteritis virus. J. Gen. Virol. 1993;74:1795–1804. - PubMed
-
- Bridgen A., Kocherhans K., Tobler K., Carvajal A., Ackermann M. Further analysis of the genome of porcine epidemic diarrhea virus. Adv. Exp. Med. Biol. 1998;101:781–786. - PubMed
-
- Britton P., Kottier S., Chen C.M., Pocock D.H., Salmon H., Aynaud J.M. The use of PCR genome mapping for characterization of TGEV strains. Adv. Exp. Med. Biol. 1994;342:29–34. - PubMed
-
- Debouck P., Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus CV777. Am. J. Vet. Res. 1980;41:219–223. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

