Defining desmosomal plakophilin-3 interactions
- PMID: 12707304
- PMCID: PMC2172904
- DOI: 10.1083/jcb.200303036
Defining desmosomal plakophilin-3 interactions
Abstract
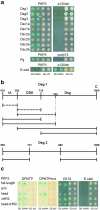
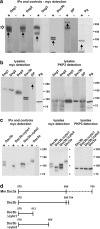
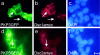
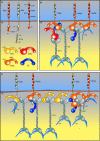
Plakophilin 3 (PKP3) is a recently described armadillo protein of the desmosomal plaque, which is synthesized in simple and stratified epithelia. We investigated the localization pattern of endogenous and exogenous PKP3 and fragments thereof. The desmosomal binding properties of PKP3 were determined using yeast two-hybrid, coimmunoprecipitation and colocalization experiments. To this end, novel mouse anti-PKP3 mAbs were generated. We found that PKP3 binds all three desmogleins, desmocollin (Dsc) 3a and -3b, and possibly also Dsc1a and -2a. As such, this is the first protein interaction ever observed with a Dsc-b isoform. Moreover, we determined that PKP3 interacts with plakoglobin, desmoplakin (DP) and the epithelial keratin 18. Evidence was found for the presence of at least two DP-PKP3 interaction sites. This finding might explain how lateral DP-PKP interactions are established in the upper layers of stratified epithelia, increasing the size of the desmosome and the number of anchoring points available for keratins. Together, these results show that PKP3, whose epithelial and epidermal desmosomal expression pattern and protein interaction repertoire are broader than those of PKP1 and -2, is a unique multiprotein binding element in the basic architecture of a vast majority of epithelial desmosomes.
Figures
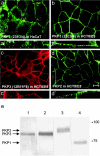


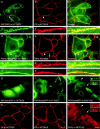
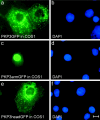
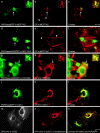

References
-
- Armstrong, D.K.B., K.E. McKenna, P.E. Purkis, K.J. Green, R.A.J. Eady, I.M. Leigh, and A.E. Hughes. 1999. Haploinsufficiency of desmoplakin causes a striate subtype of palmoplantar keratoderma. Hum. Mol. Genet. 8:143–148. - PubMed
-
- Bannon, L.J., B.L. Cabrera, M.S. Stack, and K.J. Green. 2001. Isoform-specific differences in the size of desmosomal cadherin/catenin complexes. J. Invest. Dermatol. 117:1302–1306. - PubMed
-
- Basehoar, G., and M.W. Berns. 1973. Cloning of rat kangaroo (PTK2) cells following laser microirradiation of selected mitotic chromosomes. Science. 179:1333–1334. - PubMed
-
- Bierkamp, C., K.J. McLaughlin, H. Schwarz, O. Huber, and R. Kemler. 1996. Embryonic heart and skin defects in mice lacking plakoglobin. Dev. Biol. 180:780–785. - PubMed
-
- Bonné, S., J. van Hengel, F. Nollet, P. Kools, and F. van Roy. 1999. Plakophilin-3, a novel Armadillo-like protein present in nuclei and desmosomes of epithelial cells. J. Cell Sci. 112:2265–2276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

