The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint
- PMID: 12707311
- PMCID: PMC2172906
- DOI: 10.1083/jcb.200208092
The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint
Abstract
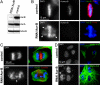
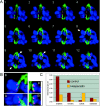
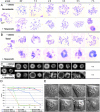
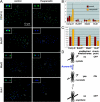
The proper segregation of sister chromatids in mitosis depends on bipolar attachment of all chromosomes to the mitotic spindle. We have identified the small molecule Hesperadin as an inhibitor of chromosome alignment and segregation. Our data imply that Hesperadin causes this phenotype by inhibiting the function of the mitotic kinase Aurora B. Mammalian cells treated with Hesperadin enter anaphase in the presence of numerous monooriented chromosomes, many of which may have both sister kinetochores attached to one spindle pole (syntelic attachment). Hesperadin also causes cells arrested by taxol or monastrol to enter anaphase within <1 h, whereas cells in nocodazole stay arrested for 3-5 h. Together, our data suggest that Aurora B is required to generate unattached kinetochores on monooriented chromosomes, which in turn could promote bipolar attachment as well as maintain checkpoint signaling.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

