Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation
- PMID: 12709013
- PMCID: PMC1782949
- DOI: 10.1046/j.1365-2567.2003.01598.x
Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation
Abstract
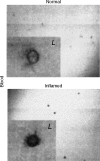
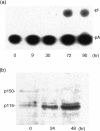
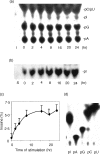
Adenosine-to-inosine (A-to-I) RNA editing is a post-transcriptional modification of pre-mRNA catalysed by an RNA-specific adenosine deaminase (ADAR). A-to-I RNA editing has been previously reported in the pre-mRNAs of brain glutamate and serotonin receptors and in lung tissue during inflammation. Here we report that systemic inflammation markedly induces inosine-containing mRNA to approximately 5% of adenosine in total mRNA. Induction was the result of up-regulation of A-to-I RNA editing as both dsRNA editing activity and ADAR1 expression were increased in the spleen, thymus and peripheral lymphocytes from endotoxin-treated mice. Up-regulation of ADAR1 was confirmed in vitro in T lymphocytes and macrophages stimulated with a variety of inflammatory mediators including tumour necrosis factor-alpha and interferon-gamma. A late induction of RNA editing was detected in concanavalin A-activated splenocytes stimulated with interleukin-2 in vitro. Taken together, these data suggest that a large number of inosine-containing mRNAs are produced during acute inflammation via up-regulation of ADAR1-mediated RNA editing. These events may affect the inflammatory and immune response through modulation of protein production.
Figures




References
-
- Yang JH, Sklar P, Axel R, Maniatis T. Editing of glutamate receptor subunit B pre-mRNA in vitro by site-specific deamination of adenosine. Nature. 1995;374:77–81. - PubMed
-
- Melcher T, Maas S, Higuchi M, Keller W, Seeburg PH. Editing of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor-B pre-mRNA in vitro reveals site-selective adenosine to inosine conversion. J Biol Chem. 1995;270:8566–70. - PubMed
-
- Rueter SM, Burns CM, Coode SA, Mookherjee P, Emeson RB. Glutamate receptor RNA editing in vitro by enzymatic conversion of adenosine to inosine. Science. 1995;267:1491–4. - PubMed
-
- Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

