The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs
- PMID: 12711670
- PMCID: PMC154230
- DOI: 10.1093/nar/gkg344
The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs
Abstract
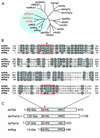
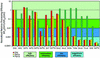
Human thymine-DNA glycosylase (TDG) is well known to excise thymine and uracil from G.T and G.U mismatches, respectively, and was therefore proposed to play a central role in the cellular defense against genetic mutation through spontaneous deamination of 5-methylcytosine and cytosine. In this study, we characterized two newly discovered orthologs of TDG, the Drosophila melanogaster Thd1p and the Schizosaccharomyces pombe Thp1p proteins, with an objective to address the function of this subfamily of uracil-DNA glycosylases from an evolutionary perspective. A systematic biochemical comparison of both enzymes with human TDG revealed a number of biologically significant facts. (i) All eukaryotic TDG orthologs have broad and species-specific substrate spectra that include a variety of damaged pyrimidine and purine bases; (ii) the common most efficiently processed substrates of all are uracil and 3,N4- ethenocytosine opposite guanine and 5-fluorouracil in any double-stranded DNA context; (iii) 5-methylcytosine and thymine derivatives are processed with an appreciable efficiency only by the human and the Drosophila enzymes; (iv) none of the proteins is able to hydrolyze a non-damaged 5'-methylcytosine opposite G; and (v) the double strand and mismatch dependency of the enzymes varies with the substrate and is not a stringent feature of this subfamily of DNA glycosylases. These findings advance our current view on the role of TDG proteins and document that they have evolved with high structural flexibility to counter a broad range of DNA base damage in accordance with the specific needs of individual species.
Figures



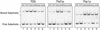

Similar articles
-
A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase.Nature. 1996 Oct 24;383(6602):735-8. doi: 10.1038/383735a0. Nature. 1996. PMID: 8878487
-
Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg:G mispair.Nucleic Acids Res. 2003 Sep 15;31(18):5399-404. doi: 10.1093/nar/gkg730. Nucleic Acids Res. 2003. PMID: 12954776 Free PMC article.
-
Repair of deaminated base damage by Schizosaccharomyces pombe thymine DNA glycosylase.DNA Repair (Amst). 2008 Dec 1;7(12):1962-72. doi: 10.1016/j.dnarep.2008.08.006. Epub 2008 Sep 25. DNA Repair (Amst). 2008. PMID: 18789404
-
Thymine DNA glycosylase.Prog Nucleic Acid Res Mol Biol. 2001;68:235-53. doi: 10.1016/s0079-6603(01)68103-0. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554300 Review.
-
Structure and function in the uracil-DNA glycosylase superfamily.Mutat Res. 2000 Aug 30;460(3-4):165-81. doi: 10.1016/s0921-8777(00)00025-2. Mutat Res. 2000. PMID: 10946227 Review.
Cited by
-
The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase.Nucleic Acids Res. 2007;35(18):6207-18. doi: 10.1093/nar/gkm678. Epub 2007 Sep 12. Nucleic Acids Res. 2007. PMID: 17855402 Free PMC article.
-
Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks.Plant Mol Biol. 2008 Aug;67(6):671-81. doi: 10.1007/s11103-008-9346-0. Epub 2008 May 21. Plant Mol Biol. 2008. PMID: 18493721
-
The cellular, developmental and population-genetic determinants of mutation-rate evolution.Genetics. 2008 Oct;180(2):933-43. doi: 10.1534/genetics.108.090456. Epub 2008 Aug 30. Genetics. 2008. PMID: 18757919 Free PMC article.
-
Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism.Nat Commun. 2016 Mar 2;7:10806. doi: 10.1038/ncomms10806. Nat Commun. 2016. PMID: 26932196 Free PMC article.
-
Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases.Acc Chem Res. 2009 Jan 20;42(1):97-106. doi: 10.1021/ar800114w. Acc Chem Res. 2009. PMID: 18837522 Free PMC article.
References
-
- Hardeland U., Bentele,M., Lettieri,T., Steinacher,R., Jiricny,J. and Schär,P. (2001) Prog. Nucleic Acids Res. Mol. Biol., 68, 235–252. - PubMed
-
- Schärer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays, 23, 270–281. - PubMed
-
- Slupphaug G., Eftedal,I., Kavli,B., Bharati,S., Helle,N.M., Haug,T., Levine,D.W. and Krokan,H.E. (1995) Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry, 34, 128–138. - PubMed
-
- Waters T.R. and Swann,P.F. (1998) Kinetics of the action of thymine DNA glycosylase. J. Biol. Chem., 273, 20007–20014. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

