Identification and characterization of transcription factor IIIA and ribosomal protein L5 from Arabidopsis thaliana
- PMID: 12711688
- PMCID: PMC154221
- DOI: 10.1093/nar/gkg335
Identification and characterization of transcription factor IIIA and ribosomal protein L5 from Arabidopsis thaliana
Abstract
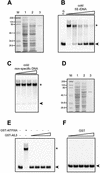
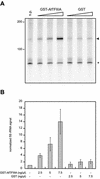
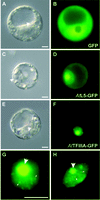
Thus far, no transcription factor IIIA (TFIIIA) from higher plants has been cloned and characterized. We have cloned and characterized TFIIIA and ribosomal protein L5 from Arabidopsis thaliana. Primary sequence comparison revealed a high divergence of AtTFIIIA and a relatively high conservation of AtL5 when compared with other organisms. The AtTFIIIA cDNA encodes a protein with nine Cys(2)-His(2)-type zinc fingers, a 23 amino acid spacer between fingers 1 and 2, a 66 amino acid spacer between fingers 4 and 5, and a 50 amino acid non-finger C-terminal tail. Aside from the amino acids required for proper zinc finger folding, AtTFIIIA is highly divergent from other known TFIIIAs. AtTFIIIA can bind 5S rDNA, as well as 5S rRNA, and efficiently stimulates the transcription of an Arabidopsis 5S rRNA gene in vitro. AtL5 identity was confirmed by demonstrating that this protein binds to 5S rRNA but not to 5S rDNA. Protoplast transient expression assays with green fluorescent protein fusion proteins revealed that AtTFIIIA is absent from the cytoplasm and concentrated at several nuclear foci including the nucleolus. AtL5 protein accumulates in the nucleus, especially in the nucleolus, and is also present in the cytoplasm.
Figures




Similar articles
-
Identification and characterization of transcription factor IIIA from Schizosaccharomyces pombe.Nucleic Acids Res. 2002 Jul 1;30(13):2772-81. doi: 10.1093/nar/gkf385. Nucleic Acids Res. 2002. PMID: 12087160 Free PMC article.
-
Molecular biology of vertebrate transcription factor IIIA: cloning and characterization of TFIIIA from channel catfish oocytes.Gene. 1997 Dec 12;203(2):103-12. doi: 10.1016/s0378-1119(97)00499-x. Gene. 1997. PMID: 9426240
-
cDNA cloning, DNA binding, and evolution of mammalian transcription factor IIIA.Gene. 2002 Jan 9;282(1-2):43-52. doi: 10.1016/s0378-1119(01)00796-x. Gene. 2002. PMID: 11814676
-
Structure, function and regulation of Transcription Factor IIIA: From Xenopus to Arabidopsis.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):274-82. doi: 10.1016/j.bbagrm.2012.10.013. Epub 2012 Nov 8. Biochim Biophys Acta. 2013. PMID: 23142779 Review.
-
Regulation of Pol I-transcribed 45S rDNA and Pol III-transcribed 5S rDNA in Arabidopsis.Plant Cell Physiol. 2012 Feb;53(2):267-76. doi: 10.1093/pcp/pcr177. Epub 2011 Dec 14. Plant Cell Physiol. 2012. PMID: 22173098 Review.
Cited by
-
Arabidopsis AN3 and OLIGOCELLULA genes link telomere maintenance mechanisms with cell division and expansion control.Plant Mol Biol. 2024 May 30;114(3):65. doi: 10.1007/s11103-024-01457-6. Plant Mol Biol. 2024. PMID: 38816532 Free PMC article.
-
Low abundant spacer 5S rRNA transcripts are frequently polyadenylated in Nicotiana.Mol Genet Genomics. 2007 Nov;278(5):565-73. doi: 10.1007/s00438-007-0273-6. Epub 2007 Aug 2. Mol Genet Genomics. 2007. PMID: 17671796
-
Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome.BMC Genomics. 2004 Jul 5;5(1):39. doi: 10.1186/1471-2164-5-39. BMC Genomics. 2004. PMID: 15236668 Free PMC article.
-
BcTFIIIA Negatively Regulates Turnip Mosaic Virus Infection through Interaction with Viral CP and VPg Proteins in Pak Choi (Brassica campestris ssp. chinensis).Genes (Basel). 2022 Jul 6;13(7):1209. doi: 10.3390/genes13071209. Genes (Basel). 2022. PMID: 35885992 Free PMC article.
-
A survey of well conserved families of C2H2 zinc-finger genes in Daphnia.BMC Genomics. 2010 Apr 30;11:276. doi: 10.1186/1471-2164-11-276. BMC Genomics. 2010. PMID: 20433734 Free PMC article.
References
-
- Bieker J.J., Martin,P.L. and Roeder,R.G. (1985) Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell, 40, 119–127. - PubMed
-
- Setzer D.R. and Brown,D.D. (1985) Formation and stability of the 5 S RNA transcription complex. J. Biol. Chem., 260, 2483–2492. - PubMed
-
- Ginsberg A.M., King,B.O. and Roeder,R.G. (1984) Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell, 39, 479–489. - PubMed
-
- Gaskins C.J., Smith,J.F., Ogilvie,M.K. and Hanas,J.S. (1992) Comparison of the sequence and structure of transcription factor IIIA from Bufo americanus and Rana pipiens. Gene, 120, 197–206. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

