Topological structure analysis of the protein-protein interaction network in budding yeast
- PMID: 12711690
- PMCID: PMC154226
- DOI: 10.1093/nar/gkg340
Topological structure analysis of the protein-protein interaction network in budding yeast
Abstract
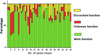
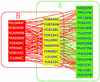
Interaction detection methods have led to the discovery of thousands of interactions between proteins, and discerning relevance within large-scale data sets is important to present-day biology. Here, a spectral method derived from graph theory was introduced to uncover hidden topological structures (i.e. quasi-cliques and quasi-bipartites) of complicated protein-protein interaction networks. Our analyses suggest that these hidden topological structures consist of biologically relevant functional groups. This result motivates a new method to predict the function of uncharacterized proteins based on the classification of known proteins within topological structures. Using this spectral analysis method, 48 quasi-cliques and six quasi-bipartites were isolated from a network involving 11,855 interactions among 2617 proteins in budding yeast, and 76 uncharacterized proteins were assigned functions.
Figures
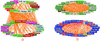
Similar articles
-
k-Partite cliques of protein interactions: A novel subgraph topology for functional coherence analysis on PPI networks.J Theor Biol. 2014 Jan 7;340:146-54. doi: 10.1016/j.jtbi.2013.09.013. Epub 2013 Sep 19. J Theor Biol. 2014. PMID: 24056214
-
Identifying protein complexes based on multiple topological structures in PPI networks.IEEE Trans Nanobioscience. 2013 Sep;12(3):165-72. doi: 10.1109/TNB.2013.2264097. Epub 2013 Aug 21. IEEE Trans Nanobioscience. 2013. PMID: 23974659
-
Topological and functional comparison of community detection algorithms in biological networks.BMC Bioinformatics. 2019 Apr 27;20(1):212. doi: 10.1186/s12859-019-2746-0. BMC Bioinformatics. 2019. PMID: 31029085 Free PMC article.
-
Protein-protein interactions: making sense of networks via graph-theoretic modeling.Bioessays. 2011 Feb;33(2):115-23. doi: 10.1002/bies.201000044. Bioessays. 2011. PMID: 21188720 Review.
-
Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast.J Cell Biol. 2002 Apr 15;157(2):199-203. doi: 10.1083/jcb.200201052. Epub 2002 Apr 15. J Cell Biol. 2002. PMID: 11956223 Free PMC article. Review.
Cited by
-
Application of approximate pattern matching in two dimensional spaces to grid layout for biochemical network maps.PLoS One. 2012;7(6):e37739. doi: 10.1371/journal.pone.0037739. Epub 2012 Jun 5. PLoS One. 2012. PMID: 22679486 Free PMC article.
-
The interactome as a tree--an attempt to visualize the protein-protein interaction network in yeast.Nucleic Acids Res. 2004 Sep 8;32(16):4804-11. doi: 10.1093/nar/gkh814. Print 2004. Nucleic Acids Res. 2004. PMID: 15356297 Free PMC article.
-
Finding local communities in protein networks.BMC Bioinformatics. 2009 Sep 18;10:297. doi: 10.1186/1471-2105-10-297. BMC Bioinformatics. 2009. PMID: 19765306 Free PMC article.
-
Topological properties of protein-protein and metabolic interaction networks of Drosophila melanogaster.Genomics Proteomics Bioinformatics. 2006 May;4(2):80-9. doi: 10.1016/S1672-0229(06)60020-X. Genomics Proteomics Bioinformatics. 2006. PMID: 16970548 Free PMC article.
-
Revealing the hidden relationship by sparse modules in complex networks with a large-scale analysis.PLoS One. 2013 Jun 10;8(6):e66020. doi: 10.1371/journal.pone.0066020. Print 2013. PLoS One. 2013. PMID: 23762457 Free PMC article.
References
-
- Fields S. (1997) The future is function. Nature Genet., 15, 325–327. - PubMed
-
- Rain J.C., Selig,L., De Reuse,H., Battaglia,V., Reverdy,C., Simon,S., Lenzen,G., Petel,F., Wojcik,J., Schachter,V., Chemama,Y., Labigne,A. and Legrain,P. (2001) The protein–protein interaction map of Helicobacter pylori. Nature, 409, 211–215. - PubMed
-
- Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. - PubMed
-
- von Mering C., Krause,R., Snel,B., Cornell,M., Oliver,S.G., Fields,S. and Bork,P. (2002) Comparative assessment of large-scale data sets of protein–protein interactions. Nature, 417, 399–403. - PubMed
-
- Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Narayan,V., Lockshon,D., Srinivasan,M., Pochart,P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

