Stimulation of human DNA polymerase epsilon by MDM2
- PMID: 12711691
- PMCID: PMC154228
- DOI: 10.1093/nar/gkg342
Stimulation of human DNA polymerase epsilon by MDM2
Abstract
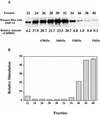
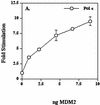
The human DNA polymerase epsilon catalytic subunit consists of a 140-kDa N-terminal domain that contains the catalytic activity and a 120-kDa C-terminal domain that binds to the other subunits and to exogenous peptides, including PCNA and MDM2. We report here that recombinant human MDM2 purified from insect cells or Escherichia coli stimulated the activity of DNA polymerase epsilon up to 10- and 40-fold, respectively, but not those of DNA polymerase beta or Klenow fragment of E.coli DNA polymerase I. Kinetic studies indicated that MDM2 increased the maximum velocity of the reaction, but did not change substrate affinities. The stimulation depended upon the interaction of the N-terminal 166 amino acid residues of MDM2 with the C-terminal domain of the full-length catalytic subunit, since the deletion of 166 amino acids from N-terminal of MDM2 or the removal of the C-terminal domain of DNA polymerase epsilon by trypsin digestion or competition for binding to it by the addition of excess C-terminal fragment eliminated the stimulation. Since DNA polymerase epsilon appears to be involved in DNA replication, recombination and repair synthesis, we suggest that MDM2 binding to DNA polymerase epsilon might be part of a reconfiguration process that allows DNA polymerase epsilon to associate with repair/recombination proteins in response to DNA damage.
Figures







Similar articles
-
MDM2 interacts with the C-terminus of the catalytic subunit of DNA polymerase epsilon.Nucleic Acids Res. 2000 Sep 15;28(18):3581-6. doi: 10.1093/nar/28.18.3581. Nucleic Acids Res. 2000. PMID: 10982879 Free PMC article.
-
Purification, cDNA cloning, and gene mapping of the small subunit of human DNA polymerase epsilon.J Biol Chem. 1997 Dec 19;272(51):32337-44. doi: 10.1074/jbc.272.51.32337. J Biol Chem. 1997. PMID: 9405441
-
Proteolysis of the human DNA polymerase epsilon catalytic subunit by caspase-3 and calpain specifically during apoptosis.Nucleic Acids Res. 2000 Nov 1;28(21):4180-8. doi: 10.1093/nar/28.21.4180. Nucleic Acids Res. 2000. PMID: 11058115 Free PMC article.
-
DNA polymerase epsilon - more than a polymerase.ScientificWorldJournal. 2003 Mar 24;3:87-104. doi: 10.1100/tsw.2003.08. ScientificWorldJournal. 2003. PMID: 12806123 Free PMC article. Review.
-
DNA polymerase epsilon: a polymerase of unusual size (and complexity).Prog Nucleic Acid Res Mol Biol. 2008;82:101-45. doi: 10.1016/S0079-6603(08)00004-4. Prog Nucleic Acid Res Mol Biol. 2008. PMID: 18929140 Free PMC article. Review. No abstract available.
Cited by
-
The potential mechanism of extracellular high mobility group box-1 protein mediated p53 expression in immune dysfunction of T lymphocytes.Oncotarget. 2017 Dec 4;8(68):112959-112971. doi: 10.18632/oncotarget.22913. eCollection 2017 Dec 22. Oncotarget. 2017. PMID: 29348880 Free PMC article.
-
Functional analysis of Drosophila DNA polymerase ε p58 subunit.Am J Cancer Res. 2013 Nov 1;3(5):478-89. eCollection 2013. Am J Cancer Res. 2013. PMID: 24224125 Free PMC article.
-
The pyrido[b]indole MDM2 inhibitor SP-141 exerts potent therapeutic effects in breast cancer models.Nat Commun. 2014 Oct 1;5:5086. doi: 10.1038/ncomms6086. Nat Commun. 2014. PMID: 25271708 Free PMC article.
-
Identification of a new class of MDM2 inhibitor that inhibits growth of orthotopic pancreatic tumors in mice.Gastroenterology. 2014 Oct;147(4):893-902.e2. doi: 10.1053/j.gastro.2014.07.001. Epub 2014 Jul 10. Gastroenterology. 2014. PMID: 25016295 Free PMC article.
-
The MDM2-p53 pathway revisited.J Biomed Res. 2013 Jul;27(4):254-71. doi: 10.7555/JBR.27.20130030. Epub 2013 Jun 6. J Biomed Res. 2013. PMID: 23885265 Free PMC article.
References
-
- Oliner J.D., Kinzler,K.W., Meltzer,P.S., George,D.L. and Vogelstein,B. (1992) Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature, 358, 80–83. - PubMed
-
- Haupt Y., Maya,R., Kazaz,A. and Oren,M. (1997) Mdm2 promotes the rapid degradation of p53. Nature, 387, 296–299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

