RNase T1 mediated base-specific cleavage and MALDI-TOF MS for high-throughput comparative sequence analysis
- PMID: 12711692
- PMCID: PMC154235
- DOI: 10.1093/nar/gng047
RNase T1 mediated base-specific cleavage and MALDI-TOF MS for high-throughput comparative sequence analysis
Abstract
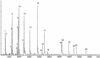
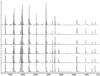
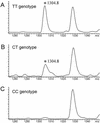
Here we devise a new method for high-throughput comparative sequence analysis. The developed protocol comprises a homogeneous in vitro transcription/RNase cleavage system with the accuracy and data acquisition speed of matrix-assisted laser desorption/ionization coupled with time-of-flight mass spectrometry (MALDI-TOF MS). In summary, the target region is PCR amplified using primers tagged with promoter sequences of T7 or SP6 RNA polymerase. Using RNase T1, the in vitro transcripts are base-specifically cleaved at every G-position. This reaction results in a characteristic pattern of fragment masses that is indicative of the original target sequence. To enable high-throughput analysis, samples are processed with automated liquid handling devices and nanoliter amounts are dispensed onto SpectroCHIP arrays for reliable and homogeneous MALDI preparation. This system enables rapid automated comparative sequence analysis for PCR products up to 1 kb in length. We demonstrate the feasibility of the devised method for analysis of single nucleotide polymorphisms (SNPs) and pathogen identification.
Figures




Similar articles
-
Rapid analysis of CpG methylation patterns using RNase T1 cleavage and MALDI-TOF.Nucleic Acids Res. 2004 Dec 2;32(21):e167. doi: 10.1093/nar/gnh165. Nucleic Acids Res. 2004. PMID: 15576674 Free PMC article.
-
Rapid and accurate characterisation of short tandem repeats by MALDI-TOF analysis of endonuclease cleaved RNA transcripts.Nucleic Acids Res. 2004 Jan 20;32(2):e16. doi: 10.1093/nar/gnh017. Nucleic Acids Res. 2004. PMID: 14734817 Free PMC article.
-
Novel mass spectrometry-based tool for genotypic identification of mycobacteria.J Clin Microbiol. 2004 Jan;42(1):339-46. doi: 10.1128/JCM.42.1.339-346.2004. J Clin Microbiol. 2004. PMID: 14715774 Free PMC article.
-
MALDI-TOF-MS analysis of protein and DNA.Neuroscientist. 2001 Feb;7(1):6-12. doi: 10.1177/107385840100700104. Neuroscientist. 2001. PMID: 11486345 Review.
-
[Analysis of polymorphic markers by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005 Apr;22(2):185-8. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005. PMID: 15793781 Review. Chinese.
Cited by
-
MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis.Mol Biotechnol. 2004 Feb;26(2):147-64. doi: 10.1385/MB:26:2:147. Mol Biotechnol. 2004. PMID: 14764940 Review.
-
High-throughput homogeneous mass cleave assay technology for the diagnosis of autosomal recessive Parkinson's disease.J Mol Diagn. 2008 May;10(3):217-24. doi: 10.2353/jmoldx.2008.070100. Epub 2008 Apr 10. J Mol Diagn. 2008. PMID: 18403612 Free PMC article.
-
Two combinatorial optimization problems for SNP discovery using base-specific cleavage and mass spectrometry.BMC Syst Biol. 2012;6 Suppl 2(Suppl 2):S5. doi: 10.1186/1752-0509-6-S2-S5. Epub 2012 Dec 12. BMC Syst Biol. 2012. PMID: 23282116 Free PMC article.
-
Microbial identification by mass cataloging.BMC Bioinformatics. 2006 Mar 8;7:117. doi: 10.1186/1471-2105-7-117. BMC Bioinformatics. 2006. PMID: 16524471 Free PMC article.
-
Rapid analysis of CpG methylation patterns using RNase T1 cleavage and MALDI-TOF.Nucleic Acids Res. 2004 Dec 2;32(21):e167. doi: 10.1093/nar/gnh165. Nucleic Acids Res. 2004. PMID: 15576674 Free PMC article.
References
-
- Blackwell J.M. (2001) Genetics and genomics in infectious disease susceptibility. Trends Mol. Med., 7, 521–526. - PubMed
-
- Chamberlain J.C. and Joubert,P.H. (2001) Opportunities and strategies for introducing pharmacogenetics into early drug development. Drug Discov. Today, 6, 569–574. - PubMed
-
- Hungnes O., Jonassen,T.O., Jonassen,C.M. and Grinde,B. (2000) Molecular epidemiology of viral infections. How sequence information helps us understand the evolution and dissemination of viruses. APMIS 2000, 108, 81–97. - PubMed
-
- Muddiman D.C., Anderson,G.A., Hofstadler,S.A. and Smith,R.D. (1997) Length and base composition of PCR-amplified nucleic acids using mass measurements from electrospray ionization mass spectrometry. Anal. Chem., 69, 1543–1549. - PubMed
-
- Null A.P., Hannis,J.C. and Muddiman,D.C. (2001) Genotyping of simple and compound short tandem repeat loci using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem., 73, 4514–4521. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

