Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress
- PMID: 12716947
- PMCID: PMC6742304
- DOI: 10.1523/JNEUROSCI.23-08-03394.2003
Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress
Abstract
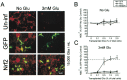
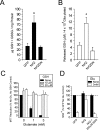
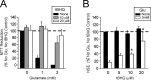
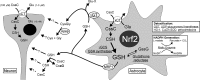
Astrocytes have a higher antioxidant potential in comparison to neurons. Pathways associated with this selective advantage include the transcriptional regulation of antioxidant enzymes via the action of the Cap'n'Collar transcription factor Nrf2 at the antioxidant response element (ARE). Here we show that Nrf2 overexpression can reengineer neurons to express this glial pathway and enhance antioxidant gene expression. However, Nrf2-mediated protection from oxidative stress is conferred primarily by glia in mixed cultures. The antioxidant properties of Nrf2-overexpressing glia are more pronounced than those of neurons, and a relatively small number of these glia (< 1% of total cell number added) could protect fully cocultured naive neurons from oxidative glutamate toxicity associated with glutathione (GSH) depletion. Microarray and biochemical analyses indicate a coordinated upregulation of enzymes involved in GSH biosynthesis (xCT cystine antiporter, gamma-glutamylcysteine synthetase, and GSH synthase), use (glutathione S-transferase and glutathione reductase), and export (multidrug resistance protein 1) with Nrf2 overexpression, leading to an increase in both media and intracellular GSH. Selective inhibition of glial GSH synthesis and the supplementation of media GSH indicated that an Nrf2-dependent increase in glial GSH synthesis was both necessary and sufficient for the protection of neurons, respectively. Neuroprotection was not limited to overexpression of Nrf2, because activation of endogenous glial Nrf2 by the small molecule ARE inducer, tert-butylhydroquinone, also protected against oxidative glutamate toxicity.
Figures

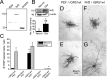
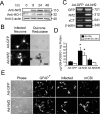
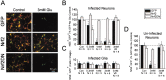
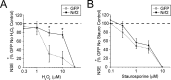
References
-
- Ahlgren-Beckendorf JA, Reising AM, Schander MA, Herdler JW, Johnson JA. Coordinate regulation of NAD(P)H:quinone oxidoreductase and glutathione S-transferases in primary cultures of rat neurons and glia: role of the antioxidant/electrophile responsive element. Glia. 1999;25:131–142. - PubMed
-
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. - PubMed
-
- Araki K, Harada M, Ueda Y, Takino T, Kuriyama K. Alteration of amino acid content of cerebrospinal fluid from patients with epilepsy. Acta Neurol Scand. 1988;78:473–479. - PubMed
-
- Bannai S. Exchange of cystine and glutamate across plasma membrane of human fibroblasts. J Biol Chem. 1986;261:2256–2263. - PubMed
-
- Bloom D, Dhakshinamoorthy S, Jaiswal AK. Site-directed mutagenesis of cysteine to serine in the DNA binding region of Nrf2 decreases its capacity to upregulate antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2002;21:2191–2200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
