Ligand-induced changes in dynamics in the RT loop of the C-terminal SH3 domain of Sem-5 indicate cooperative conformational coupling
- PMID: 12717021
- PMCID: PMC2323869
- DOI: 10.1110/ps.0238003
Ligand-induced changes in dynamics in the RT loop of the C-terminal SH3 domain of Sem-5 indicate cooperative conformational coupling
Abstract
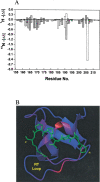
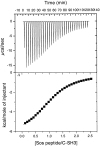
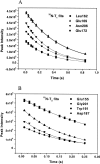
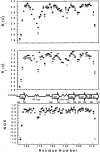
We report the effects of peptide binding on the (15)N relaxation rates and chemical shifts of the C-SH3 of Sem-5. (15)N spin-lattice relaxation time (T(1)), spin-spin relaxation time (T(2)), and ((1)H)-(15)N NOE were obtained from heteronuclear 2D NMR experiments. These parameters were then analyzed using the Lipari-Szabo model free formalism to obtain parameters that describe the internal motions of the protein. High-order parameters (S(2) > 0.8) are found in elements of regular secondary structure, whereas some residues in the loop regions show relatively low-order parameters, notably the RT loop. Peptide binding is characterized by a significant decrease in the (15)N relaxation in the RT loop. Concomitant with the change in dynamics is a cooperative change in chemical shifts. The agreement between the binding constants calculated from chemical shift differences and that obtained from ITC indicates that the binding of Sem-5 C-SH3 to its putative peptide ligand is coupled to a cooperative conformational change in which a portion of the binding site undergoes a significant reduction in conformational heterogeneity.
Figures





Similar articles
-
Directed discovery of bivalent peptide ligands to an SH3 domain.Protein Sci. 2004 Mar;13(3):626-32. doi: 10.1110/ps.03470504. Protein Sci. 2004. PMID: 14978303 Free PMC article.
-
Backbone dynamics of the C-terminal SH2 domain of the p85alpha subunit of phosphoinositide 3-kinase: effect of phosphotyrosine-peptide binding and characterization of slow conformational exchange processes.J Mol Biol. 2000 Jun 9;299(3):771-88. doi: 10.1006/jmbi.2000.3760. J Mol Biol. 2000. PMID: 10835283
-
Determination of the solution structure of the SH3 domain of human p56 Lck tyrosine kinase.J Biomol NMR. 1996 Sep;8(2):105-22. doi: 10.1007/BF00211158. J Biomol NMR. 1996. PMID: 8914270
-
Characterization of enzyme motions by solution NMR relaxation dispersion.Acc Chem Res. 2008 Feb;41(2):214-21. doi: 10.1021/ar700132n. Epub 2008 Feb 19. Acc Chem Res. 2008. PMID: 18281945 Review.
-
Enzyme dynamics from NMR spectroscopy.Acc Chem Res. 2015 Feb 17;48(2):457-65. doi: 10.1021/ar500340a. Epub 2015 Jan 9. Acc Chem Res. 2015. PMID: 25574774 Free PMC article. Review.
Cited by
-
A statistical analysis of the PPII propensity of amino acid guests in proline-rich peptides.Biophys J. 2011 Feb 16;100(4):1083-93. doi: 10.1016/j.bpj.2010.12.3742. Biophys J. 2011. PMID: 21320454 Free PMC article.
-
Resolution of Site-Specific Conformational Heterogeneity in Proline-Rich Molecular Recognition by Src Homology 3 Domains.J Am Chem Soc. 2016 Feb 3;138(4):1130-3. doi: 10.1021/jacs.5b11999. Epub 2016 Jan 25. J Am Chem Soc. 2016. PMID: 26784847 Free PMC article.
-
Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery.Pharmacol Rev. 2010 Jun;62(2):265-304. doi: 10.1124/pr.108.000992. Epub 2010 Apr 14. Pharmacol Rev. 2010. PMID: 20392808 Free PMC article. Review.
-
Rapid Quantification of Protein-Ligand Binding via 19F NMR Lineshape Analysis.Biophys J. 2020 May 19;118(10):2537-2548. doi: 10.1016/j.bpj.2020.03.031. Epub 2020 Apr 15. Biophys J. 2020. PMID: 32348722 Free PMC article.
-
SH3 domains of Grb2 adaptor bind to PXpsiPXR motifs within the Sos1 nucleotide exchange factor in a discriminate manner.Biochemistry. 2009 May 19;48(19):4074-85. doi: 10.1021/bi802291y. Biochemistry. 2009. PMID: 19323566 Free PMC article.
References
-
- Abragam, A. 1961. Principles of nuclear magnetism. Clarendon Press, Oxford, UK.
-
- Akke, M., Bruschweiler, R., and Palmer III, A.G. 1993. NMR order parameters and free-energy: An analytical approach and its application to cooperative Ca2+ binding by calbindin-d(9k). J. Am. Chem. Soc. 115 9832–9833.
-
- Alexandrescu, A.T., Rathgeb-Szabo, K., Rumpel, K., Jahnke, W., Schulthess, T., and Kammerer, R.A. 1998. 15N backbone dynamics of the S-peptide from ribonuclease A in its free and S-protein bound forms: Toward a site-specific analysis of entropy changes upon folding. Protein Sci. 7 389–402. - PMC - PubMed
-
- Booker, G.W., Gout, I., Downing, A.K., Driscoll, P.C., Boyd, J., Waterfield, M.D., and Campbell, I.D. 1993. Solution structure and ligand-binding site of the SH3 domain of the p85α subunit of phosphatidylinositol 3-kinase. Cell 73 813–822. - PubMed
-
- Buday, L. 1999. Membrane-targeting of signalling molecules by SH2/SH3 domain-containing adaptor proteins. Biochim. Biophys. Acta 1422 187–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

