Ligand-induced changes in dynamics in the RT loop of the C-terminal SH3 domain of Sem-5 indicate cooperative conformational coupling
- PMID: 12717021
- PMCID: PMC2323869
- DOI: 10.1110/ps.0238003
Ligand-induced changes in dynamics in the RT loop of the C-terminal SH3 domain of Sem-5 indicate cooperative conformational coupling
Abstract
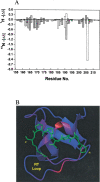
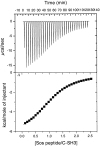
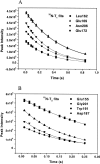
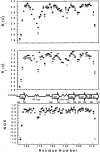
We report the effects of peptide binding on the (15)N relaxation rates and chemical shifts of the C-SH3 of Sem-5. (15)N spin-lattice relaxation time (T(1)), spin-spin relaxation time (T(2)), and ((1)H)-(15)N NOE were obtained from heteronuclear 2D NMR experiments. These parameters were then analyzed using the Lipari-Szabo model free formalism to obtain parameters that describe the internal motions of the protein. High-order parameters (S(2) > 0.8) are found in elements of regular secondary structure, whereas some residues in the loop regions show relatively low-order parameters, notably the RT loop. Peptide binding is characterized by a significant decrease in the (15)N relaxation in the RT loop. Concomitant with the change in dynamics is a cooperative change in chemical shifts. The agreement between the binding constants calculated from chemical shift differences and that obtained from ITC indicates that the binding of Sem-5 C-SH3 to its putative peptide ligand is coupled to a cooperative conformational change in which a portion of the binding site undergoes a significant reduction in conformational heterogeneity.
Figures





References
-
- Abragam, A. 1961. Principles of nuclear magnetism. Clarendon Press, Oxford, UK.
-
- Akke, M., Bruschweiler, R., and Palmer III, A.G. 1993. NMR order parameters and free-energy: An analytical approach and its application to cooperative Ca2+ binding by calbindin-d(9k). J. Am. Chem. Soc. 115 9832–9833.
-
- Alexandrescu, A.T., Rathgeb-Szabo, K., Rumpel, K., Jahnke, W., Schulthess, T., and Kammerer, R.A. 1998. 15N backbone dynamics of the S-peptide from ribonuclease A in its free and S-protein bound forms: Toward a site-specific analysis of entropy changes upon folding. Protein Sci. 7 389–402. - PMC - PubMed
-
- Booker, G.W., Gout, I., Downing, A.K., Driscoll, P.C., Boyd, J., Waterfield, M.D., and Campbell, I.D. 1993. Solution structure and ligand-binding site of the SH3 domain of the p85α subunit of phosphatidylinositol 3-kinase. Cell 73 813–822. - PubMed
-
- Buday, L. 1999. Membrane-targeting of signalling molecules by SH2/SH3 domain-containing adaptor proteins. Biochim. Biophys. Acta 1422 187–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

