Can correct protein models be identified?
- PMID: 12717029
- PMCID: PMC2323877
- DOI: 10.1110/ps.0236803
Can correct protein models be identified?
Abstract
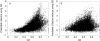
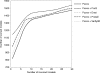
The ability to separate correct models of protein structures from less correct models is of the greatest importance for protein structure prediction methods. Several studies have examined the ability of different types of energy function to detect the native, or native-like, protein structure from a large set of decoys. In contrast to earlier studies, we examine here the ability to detect models that only show limited structural similarity to the native structure. These correct models are defined by the existence of a fragment that shows significant similarity between this model and the native structure. It has been shown that the existence of such fragments is useful for comparing the performance between different fold recognition methods and that this performance correlates well with performance in fold recognition. We have developed ProQ, a neural-network-based method to predict the quality of a protein model that extracts structural features, such as frequency of atom-atom contacts, and predicts the quality of a model, as measured either by LGscore or MaxSub. We show that ProQ performs at least as well as other measures when identifying the native structure and is better at the detection of correct models. This performance is maintained over several different test sets. ProQ can also be combined with the Pcons fold recognition predictor (Pmodeller) to increase its performance, with the main advantage being the elimination of a few high-scoring incorrect models. Pmodeller was successful in CASP5 and results from the latest LiveBench, LiveBench-6, indicating that Pmodeller has a higher specificity than Pcons alone.
Figures
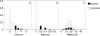

References
-
- Bauer, A. and Beyer, A. 1994. An improved pair potential to recognize native protein folds. Proteins 18 254–261. - PubMed
-
- Bishop, C.M. 1995. Neural networks for pattern recognition. Oxford University Press, Oxford, UK.
-
- Bowie, J.U., Lüthy, R., and Eisenberg, D. 1991. A method to identify protein sequence that fold into a known three-dimensional structure. Science 253 164–170. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

