Comparison of the TIM and TOM channel activities of the mitochondrial protein import complexes
- PMID: 12719229
- PMCID: PMC1302860
- DOI: 10.1016/S0006-3495(03)70024-1
Comparison of the TIM and TOM channel activities of the mitochondrial protein import complexes
Abstract
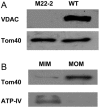
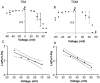
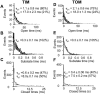
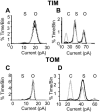
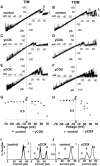
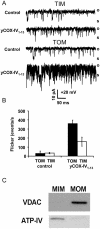
Water-filled channels are central to the process of translocating proteins since they provide aqueous pathways through the hydrophobic environment of membranes. The Tom and Tim complexes translocate precursors across the mitochondrial outer and inner membranes, respectively, and contain channels referred to as TOM and TIM (previously called PSC and MCC). In this study, little differences were revealed from a direct comparison of the single channel properties of the TOM and TIM channels of yeast mitochondria. As they perform similar functions in translocating proteins across membranes, it is not surprising that both channels are high conductance, voltage-dependent channels that are slightly cation selective. Reconstituted TIM and TOM channel activities are not modified by deletion of the outer membrane channel VDAC, but are similarly affected by signal sequence peptides.
Figures


Similar articles
-
MCC and PSC, the putative protein import channels of mitochondria.J Bioenerg Biomembr. 2000 Feb;32(1):47-54. doi: 10.1023/a:1005560328334. J Bioenerg Biomembr. 2000. PMID: 11768761 Review.
-
Tim22, the essential core of the mitochondrial protein insertion complex, forms a voltage-activated and signal-gated channel.Mol Cell. 2002 Feb;9(2):363-73. doi: 10.1016/s1097-2765(02)00446-x. Mol Cell. 2002. PMID: 11864609
-
Multiple conductance channel activity of wild-type and voltage-dependent anion-selective channel (VDAC)-less yeast mitochondria.Biophys J. 1995 Jun;68(6):2299-309. doi: 10.1016/S0006-3495(95)80412-1. Biophys J. 1995. PMID: 7544166 Free PMC article.
-
Mouse VDAC isoforms expressed in yeast: channel properties and their roles in mitochondrial outer membrane permeability.J Membr Biol. 1999 Jul 15;170(2):89-102. doi: 10.1007/s002329900540. J Membr Biol. 1999. PMID: 10430654
-
Targeting and insertion of nuclear-encoded preproteins into the mitochondrial outer membrane.Bioessays. 2000 Apr;22(4):364-71. doi: 10.1002/(SICI)1521-1878(200004)22:4<364::AID-BIES6>3.0.CO;2-N. Bioessays. 2000. PMID: 10723033 Review.
Cited by
-
A disulfide bond in the TIM23 complex is crucial for voltage gating and mitochondrial protein import.J Cell Biol. 2016 Aug 15;214(4):417-31. doi: 10.1083/jcb.201602074. Epub 2016 Aug 8. J Cell Biol. 2016. PMID: 27502485 Free PMC article.
-
A fluorescence assay for peptide translocation into mitochondria.Anal Biochem. 2007 Mar 1;362(1):76-82. doi: 10.1016/j.ab.2006.12.015. Epub 2007 Jan 22. Anal Biochem. 2007. PMID: 17240346 Free PMC article.
-
Excursion of a single polypeptide into a protein pore: simple physics, but complicated biology.Eur Biophys J. 2008 Jul;37(6):913-25. doi: 10.1007/s00249-008-0309-9. Epub 2008 Mar 27. Eur Biophys J. 2008. PMID: 18368402
-
CALHM2 is a mitochondrial protein import channel that regulates fatty acid metabolism.Res Sq [Preprint]. 2024 Sep 13:rs.3.rs-4985689. doi: 10.21203/rs.3.rs-4985689/v1. Res Sq. 2024. PMID: 39315269 Free PMC article. Preprint.
-
Proapoptotic N-truncated BCL-xL protein activates endogenous mitochondrial channels in living synaptic terminals.Proc Natl Acad Sci U S A. 2004 Sep 14;101(37):13590-5. doi: 10.1073/pnas.0401372101. Epub 2004 Sep 1. Proc Natl Acad Sci U S A. 2004. PMID: 15342906 Free PMC article.
References
-
- Bauer, M. F., S. Hofmann, W. Neupert, and M. Brunner. 2000. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 10:25–31. - PubMed
-
- Blachly-Dyson, E., S. Peng, M. Colombini, and M. Forte. 1990. Selectivity changes in site-directed mutants of the VDAC ion channel: Structural implications. Science. 247:1233–1236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

