The initiation-elongation transition: lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at +8/+9 and absent by +23
- PMID: 12719526
- PMCID: PMC156264
- DOI: 10.1073/pnas.1037057100
The initiation-elongation transition: lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at +8/+9 and absent by +23
Abstract
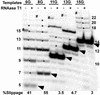
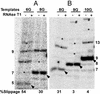
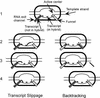
RNA polymerase II transcription complexes stalled shortly after initiation over a repetitive segment of the template can undergo efficient transcript slippage, during which the 3' end of the RNA slides upstream and then re-pairs with the template, allowing transcription to continue. In the present study, we have used transcript slippage as an assay to identify possible structural transitions that occur as the polymerase passes from the initiation to the elongation phase of transcription. We reasoned that transcript slippage would not occur in fully processive complexes. We constructed a series of templates that allowed us to stall RNA polymerase II after the synthesis of a repetitive sequence (5'-CUCUCU-3') at varying distances downstream of +1. We found that polymerase must synthesize at least a 23-nt RNA to attain resistance to transcript slippage. The ability to undergo slippage was lost in two discrete steps, suggestive of two distinct transitions. The first transition is the formation of the 8- to 9-bp mature RNA-DNA hybrid, when slippage abruptly dropped by 10-fold. However, easily detectable slippage continued until 14 more bonds were made. Thus, although the transcript becomes tightly constrained within the transcription complex once the hybrid reaches its final length, much more RNA synthesis is required before the RNA is no longer able to slip upstream along the template. This last point may reflect an important stabilizing role for the interaction of the polymerase with the transcript well upstream of the RNA-DNA hybrid.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

