Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant
- PMID: 12719547
- PMCID: PMC154014
- DOI: 10.1128/jvi.77.10.5547-5556.2003
Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant
Abstract
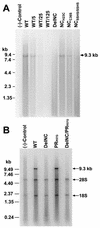
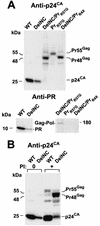
The nucleocapsid (NC) region of human immunodeficiency virus type 1 (HIV-1) Gag is required for specific genomic RNA packaging. To determine if NC is absolutely required for virion formation, we deleted all but seven amino acids from NC in a full-length NL4-3 proviral clone. This construct, DelNC, produced approximately four- to sixfold fewer virions than did the wild type, and these virions were noninfectious (less than 10(-6) relative to the wild type) and severely genomic RNA deficient. Immunoblot and high-pressure liquid chromatography analyses showed that all of the mature Gag proteins except NC were present in the mutant virion preparations, although there was a modest decrease in Gag processing. DelNC virions had lower densities and were more heterogeneous than wild-type particles, consistent with a defect in the interaction assembly or I domain. Electron microscopy showed that the DelNC virions displayed a variety of aberrant morphological forms. Inactivating the protease activity of DelNC by mutation or protease inhibitor treatment restored virion production to wild-type levels. DelNC-protease mutants formed immature-appearing particles that were as dense as wild-type virions without incorporating genomic RNA. Therefore, protease activity combined with the absence of NC causes the defect in DelNC virion production, suggesting that premature processing of Gag during assembly causes this effect. These results show that HIV-1 can form particles efficiently without NC.
Figures



References
-
- Berg, J. M., and Y. Shi. 1996. The galvanization of biology: a growing appreciation for the roles of zinc. Science 271:1081-1085. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

