Reovirus sigma NS and mu NS proteins form cytoplasmic inclusion structures in the absence of viral infection
- PMID: 12719587
- PMCID: PMC154006
- DOI: 10.1128/jvi.77.10.5948-5963.2003
Reovirus sigma NS and mu NS proteins form cytoplasmic inclusion structures in the absence of viral infection
Abstract
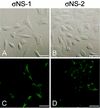
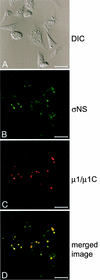
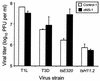
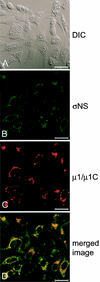
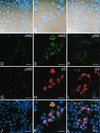
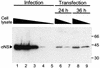
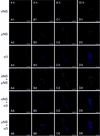
Reovirus replication occurs in the cytoplasm of infected cells and culminates in the formation of crystalline arrays of progeny virions within viral inclusions. Two viral nonstructural proteins, sigma NS and micro NS, and structural protein sigma 3 form protein-RNA complexes early in reovirus infection. To better understand the minimal requirements of viral inclusion formation, we expressed sigma NS, mu NS, and sigma 3 alone and in combination in the absence of viral infection. In contrast to its concentration in inclusion structures during reovirus replication, sigma NS expressed in cells in the absence of infection is distributed diffusely throughout the cytoplasm and does not form structures that resemble viral inclusions. Expressed sigma NS is functional as it complements the defect in temperature-sensitive, sigma NS-mutant virus tsE320. In both transfected and infected cells, mu NS is found in punctate cytoplasmic structures and sigma 3 is distributed diffusely in the cytoplasm and the nucleus. The subcellular localization of mu NS and sigma 3 is not altered when the proteins are expressed together or with sigma NS. However, when expressed with micro NS, sigma NS colocalizes with mu NS to punctate structures similar in morphology to inclusion structures observed early in viral replication. During reovirus infection, both sigma NS and mu NS are detectable 4 h after adsorption and colocalize to punctate structures throughout the viral life cycle. In concordance with these results, sigma NS interacts with mu NS in a yeast two-hybrid assay and by coimmunoprecipitation analysis. These data suggest that sigma NS and mu NS are the minimal viral components required to form inclusions, which then recruit other reovirus proteins and RNA to initiate viral genome replication.
Figures




References
-
- Antczak, J. B., and W. K. Joklik. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 187:760-776. - PubMed
-
- Astell, C., S. C. Silverstein, D. H. Levin, and G. Acs. 1972. Regulation of the reovirus RNA transcriptase by a viral capsomere protein. Virology 48:648-654. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

