Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding
- PMID: 12724402
- PMCID: PMC154245
- DOI: 10.1128/MCB.23.10.3427-3441.2003
Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding
Erratum in
- Mol Cell Biol. 2004 Jun;24(11):5088
Abstract
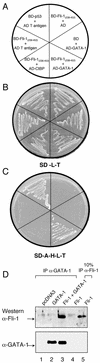
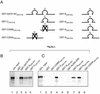
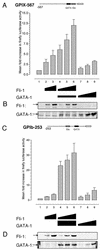
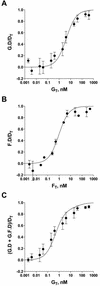
Friend leukemia integration 1 (Fli-1) is a member of the Ets family of transcriptional activators that has been shown to be an important regulator during megakaryocytic differentiation. We undertook a two-hybrid screen of a K562 cDNA library to identify transcription factors that interacted with Fli-1 and were potential regulators of megakaryocyte development. Here we report the physical interaction of Fli-1 with GATA-1, a well-characterized, zinc finger transcription factor critical for both erythroid and megakaryocytic differentiation. We map the minimal domains required for the interaction and show that the zinc fingers of GATA-1 interact with the Ets domain of Fli-1. GATA-1 has previously been shown to interact with the Ets domain of the Fli-1-related protein PU.1, and the two proteins appear to inhibit each other's activity. In contrast, we demonstrate that GATA-1 and Fli-1 synergistically activate the megakaryocyte-specific promoters GPIX and GPIbalpha in transient transfections. Quantitative electrophoretic mobility shift assays using oligonucleotides derived from the GPIX promoter containing Ets and GATA binding motifs reveal that Fli-1 and GATA-1 exhibit cooperative DNA binding in which the binding of GATA-1 to DNA is increased approximately 26-fold in the presence of Fli-1 (from 4.2 to 0.16 nM), providing a mechanism for the observed transcriptional synergy. To test the effect on endogenous genes, we stably overexpressed Fli-1 in K562 cells, a line rich in GATA-1. Overexpression of Fli-1 induced the expression of the endogenous GPIX and GPIbalpha genes as measured by Northern blot and fluorescence-activated cell sorter analysis. This work suggests that Fli-1 and GATA-1 work together to activate the expression of genes associated with the terminal differentiation of megakaryocytes.
Figures





References
-
- Athanasiou, M., P. A. Clausen, G. J. Mavrothalassitis, X. K. Zhang, D. K. Watson, and D. G. Blair. 1996. Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell Growth Differ. 7:1525-1534. - PubMed
-
- Athanasiou, M., G. Mavrothalassitis, L. Sun-Hoffman, and D. G. Blair. 2000. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia 14:439-445. - PubMed
-
- Bastian, L. S., B. A. Kwiatkowski, J. Breininger, S. Danner, and G. Roth. 1999. Regulation of the megakaryocytic glycoprotein IX promoter by the oncogenic Ets transcription factor Fli-1. Blood 93:2637-2644. - PubMed
-
- Behre, G., L. T. Smith, and D. G. Tenen. 1999. Use of a promoterless Renilla luciferase vector as an internal control plasmid for transient co-transfection assays of Ras-mediated transcription activation. BioTechniques 26:24-26, 28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
