The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase
- PMID: 12724538
- PMCID: PMC153720
- DOI: 10.1105/tpc.010827
The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase
Abstract
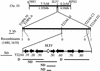
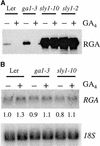
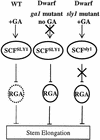
The Arabidopsis SLY1 (SLEEPY1) gene positively regulates gibberellin (GA) signaling. Positional cloning of SLY1 revealed that it encodes a putative F-box protein. This result suggests that SLY1 is the F-box subunit of an SCF E3 ubiquitin ligase that regulates GA responses. The DELLA domain protein RGA (repressor of ga1-3) is a repressor of GA response that appears to undergo GA-stimulated protein degradation. RGA is a potential substrate of SLY1, because sly1 mutations cause a significant increase in RGA protein accumulation even after GA treatment. This result suggests SCF(SLY1)-targeted degradation of RGA through the 26S proteasome pathway. Further support for this model is provided by the observation that an rga null allele partially suppresses the sly1-10 mutant phenotype. The predicted SLY1 amino acid sequence is highly conserved among plants, indicating a key role in GA response.
Figures




References
-
- Allan, R.E. (1986). Agronomic comparison among wheat lines nearly isogenic for three reduced-height genes. Crop Sci. 26, 707–710.
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- An, Y.Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

