A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae
- PMID: 12727869
- PMCID: PMC156083
- DOI: 10.1093/emboj/cdg211
A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae
Abstract
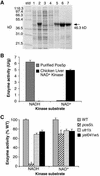
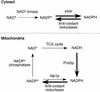
Mitochondria require NADPH for anti-oxidant protection and for specific biosynthetic pathways. However, the sources of mitochondrial NADPH and the mechanisms of maintaining mitochondrial redox balance are not well understood. We show here that in Saccharomyces cerevisiae, mitochondrial NADPH is largely provided by the product of the POS5 gene. We identified POS5 in a S.cerevisiae genetic screen for hyperoxia-sensitive mutants, or cells that cannot survive in 100% oxygen. POS5 encodes a protein that is homologous to NAD(+) and NADH kinases, and we show here that recombinant Pos5p has NADH kinase activity. Pos5p is localized to the mitochondrial matrix of yeast and appears to be important for several NADPH-requiring processes in the mitochondria, including resistance to a broad range of oxidative stress conditions, arginine biosynthesis and mitochondrial iron homeostasis. Pos5p represents the first member of the NAD(H) kinase family that has been identified as an important anti-oxidant factor and key source of the cellular reductant NADPH.
Figures





References
-
- Apps D.K. (1970) The NAD kinases of Saccharomyces cerevisiae. Eur. J. Biochem., 13, 223–230. - PubMed
-
- Becker D.M. and Guarente,L. (1991) High-efficiency transformation of yeast by electroporation. Methods Enzymol., 194, 182–187. - PubMed
-
- Bernofsky C. and Utter,M.F. (1968) Interconversions of mitochondrial pyridine nucleotides. Science, 159, 1362–1363. - PubMed
-
- Boveris A. and Cadenas,E. (1982) Production of superoxide radicals and hydrogen peroxide in mitochondria. In Oberley,L. (ed.), Superoxide Dismutase. CRC Press, Boca Raton, FL, Vol. II, pp. 15–30.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

